Mutations in the histone fold domain of the TAF12 gene show synthetic lethality with the TAF1 gene lacking the TAF N-terminal domain (TAND) by different mechanisms from those in the SPT15 gene encoding the TATA box-binding protein (TBP)
- PMID: 12582246
- PMCID: PMC150217
- DOI: 10.1093/nar/gkg180
Mutations in the histone fold domain of the TAF12 gene show synthetic lethality with the TAF1 gene lacking the TAF N-terminal domain (TAND) by different mechanisms from those in the SPT15 gene encoding the TATA box-binding protein (TBP)
Abstract
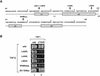
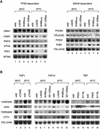
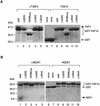
The general transcription factor TFIID, composed of the TATA box-binding protein (TBP) and 14 TBP-associated factors (TAFs), is important for both basal and regulated transcription by RNA polymerase II. Although it is well known that the TAF N-terminal domain (TAND) at the amino-terminus of the TAF1 protein binds to TBP and thereby inhibits TBP function in vitro, the physiological role of this domain remains obscure. In our previous study, we screened for mutations that cause lethality when co-expressed with the TAF1 gene lacking TAND (taf1-DeltaTAND) and identified two DeltaTAND synthetic lethal (nsl) mutations as those in the SPT15 gene encoding TBP. In this study we isolated another nsl mutation in the same screen and identified it to be a mutation in the histone fold domain (HFD) of the TAF12 gene. Several other HFD mutations of this gene also exhibit nsl phenotypes, and all of them are more or less impaired in transcriptional activation in vivo. Interestingly, a set of genes affected in the taf1-DeltaTAND mutant is similarly affected in the taf12 HFD mutants but not in the nsl mutants of TBP. Therefore, we discovered that the nsl mutations of these two genes cause lethality in the taf1-DeltaTAND mutant by different mechanisms.
Figures

Similar articles
-
Autonomous function of the amino-terminal inhibitory domain of TAF1 in transcriptional regulation.Mol Cell Biol. 2004 Apr;24(8):3089-99. doi: 10.1128/MCB.24.8.3089-3099.2004. Mol Cell Biol. 2004. PMID: 15060133 Free PMC article.
-
Mutations in the TATA-binding protein, affecting transcriptional activation, show synthetic lethality with the TAF145 gene lacking the TAF N-terminal domain in Saccharomyces cerevisiae.J Biol Chem. 2001 Jan 5;276(1):395-405. doi: 10.1074/jbc.M008208200. J Biol Chem. 2001. PMID: 11035037
-
Saccharomyces cerevisiae HMO1 interacts with TFIID and participates in start site selection by RNA polymerase II.Nucleic Acids Res. 2008 Mar;36(4):1343-57. doi: 10.1093/nar/gkm1068. Epub 2008 Jan 10. Nucleic Acids Res. 2008. PMID: 18187511 Free PMC article.
-
Roles for BTAF1 and Mot1p in dynamics of TATA-binding protein and regulation of RNA polymerase II transcription.Gene. 2003 Oct 2;315:1-13. doi: 10.1016/s0378-1119(03)00714-5. Gene. 2003. PMID: 14557059 Review.
-
Architecture of the multi-functional SAGA complex and the molecular mechanism of holding TBP.FEBS J. 2021 May;288(10):3135-3147. doi: 10.1111/febs.15563. Epub 2020 Sep 29. FEBS J. 2021. PMID: 32946670 Review.
Cited by
-
Mapping key functional sites within yeast TFIID.EMBO J. 2004 Feb 25;23(4):719-27. doi: 10.1038/sj.emboj.7600111. Epub 2004 Feb 12. EMBO J. 2004. PMID: 14765106 Free PMC article.
-
Mutation burden of rare variants in schizophrenia candidate genes.PLoS One. 2015 Jun 3;10(6):e0128988. doi: 10.1371/journal.pone.0128988. eCollection 2015. PLoS One. 2015. PMID: 26039597 Free PMC article.
-
TFIID dependency of steady-state mRNA transcription altered epigenetically by simultaneous functional loss of Taf1 and Spt3 is Hsp104-dependent.PLoS One. 2023 Feb 9;18(2):e0281233. doi: 10.1371/journal.pone.0281233. eCollection 2023. PLoS One. 2023. PMID: 36757926 Free PMC article.
-
Autonomous function of the amino-terminal inhibitory domain of TAF1 in transcriptional regulation.Mol Cell Biol. 2004 Apr;24(8):3089-99. doi: 10.1128/MCB.24.8.3089-3099.2004. Mol Cell Biol. 2004. PMID: 15060133 Free PMC article.
References
-
- Roeder R.G. (1998) Role of general and gene-specific cofactors in the regulation of eukaryotic transcription. Cold Spring Harb. Symp. Quant. Biol., 63, 201–218. - PubMed
-
- Naar A.M., Lemon,B.D. and Tjian,R. (2001) Transcriptional coactivator complexes. Annu. Rev. Biochem., 70, 475–501. - PubMed
-
- Narlikar G.J., Fan,H.Y. and Kingston,R.E. (2002) Cooperation between complexes that regulate chromatin structure and transcription. Cell, 108, 475–487. - PubMed
-
- Woychik N.A. and Hampsey,M. (2002) The RNA polymerase II machinery: structure illuminates function. Cell, 108, 453–463. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

