Inhibition of human papilloma virus E2 DNA binding protein by covalently linked polyamides
- PMID: 12582248
- PMCID: PMC150225
- DOI: 10.1093/nar/gkg206
Inhibition of human papilloma virus E2 DNA binding protein by covalently linked polyamides
Abstract
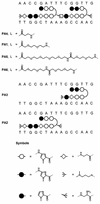
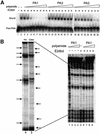
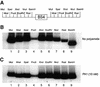
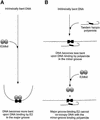
Polyamides are a class of heterocyclic small molecules with the potential of controlling gene expression by binding to the minor groove of DNA in a sequence-specific manner. To evaluate the feasibility of this class of compounds as antiviral therapeutics, molecules were designed to essential sequence elements occurring numerous times in the HPV genome. This sequence element is bound by a virus-encoded transcription and replication factor E2, which binds to a 12 bp recognition site as a homodimeric protein. Here, we take advantage of polyamide:DNA and E2:DNA co-crystal structural information and advances in polyamide synthetic chemistry to design tandem hairpin polyamides that are capable of displacing the major groove-binding E2 homodimer from its DNA binding site. The binding of tandem hairpin polyamides and the E2 DNA binding protein to the DNA site is mutually exclusive even though the two ligands occupy opposite faces of the DNA double helix. We show with circular permutation studies that the tandem hairpin polyamide prevents the intrinsic bending of the E2 DNA site important for binding of the protein. Taken together, these results illustrate the feasibility of inhibiting the binding of homodimeric, major groove-binding transcription factors by altering the local DNA geometry using minor groove-binding tandem hairpin polyamides.
Figures



Similar articles
-
Combinatorial determination of sequence specificity for nanomolar DNA-binding hairpin polyamides.Biochemistry. 2003 Jun 10;42(22):6891-903. doi: 10.1021/bi027373s. Biochemistry. 2003. PMID: 12779344
-
Inhibition of major-groove-binding proteins by pyrrole-imidazole polyamides with an Arg-Pro-Arg positive patch.Chem Biol. 1998 Mar;5(3):119-33. doi: 10.1016/s1074-5521(98)90057-6. Chem Biol. 1998. PMID: 9545429
-
DNA structure and flexibility in the sequence-specific binding of papillomavirus E2 proteins.J Mol Biol. 1998 Mar 6;276(4):809-18. doi: 10.1006/jmbi.1997.1578. J Mol Biol. 1998. PMID: 9500925
-
Recognition of the DNA minor groove by pyrrole-imidazole polyamides.Curr Opin Struct Biol. 2003 Jun;13(3):284-99. doi: 10.1016/s0959-440x(03)00081-2. Curr Opin Struct Biol. 2003. PMID: 12831879 Review.
-
Biology of N-methylpyrrole-N-methylimidazole hairpin polyamide.Biol Pharm Bull. 2004 Apr;27(4):468-74. doi: 10.1248/bpb.27.468. Biol Pharm Bull. 2004. PMID: 15056849 Review.
Cited by
-
Inhibition of heat shock transcription factor binding by a linear polyamide binding in an unusual 1:1 mode.Chembiochem. 2012 Jan 2;13(1):97-104. doi: 10.1002/cbic.201100524. Epub 2011 Dec 1. Chembiochem. 2012. PMID: 22134972 Free PMC article.
-
HPV episome levels are potently decreased by pyrrole-imidazole polyamides.Antiviral Res. 2011 Aug;91(2):177-86. doi: 10.1016/j.antiviral.2011.05.014. Epub 2011 Jun 2. Antiviral Res. 2011. PMID: 21669229 Free PMC article.
-
Modulating DNA by polyamides to regulate transcription factor PU.1-DNA binding interactions.Biochimie. 2019 Dec;167:1-11. doi: 10.1016/j.biochi.2019.08.009. Epub 2019 Aug 21. Biochimie. 2019. PMID: 31445072 Free PMC article.
-
The Reservoir of Persistent Human Papillomavirus Infection; Strategies for Elimination Using Anti-Viral Therapies.Viruses. 2022 Jan 22;14(2):214. doi: 10.3390/v14020214. Viruses. 2022. PMID: 35215808 Free PMC article. Review.
-
New Frontiers in Druggability.J Med Chem. 2015 Dec 10;58(23):9063-88. doi: 10.1021/acs.jmedchem.5b00586. Epub 2015 Aug 11. J Med Chem. 2015. PMID: 26230724 Free PMC article.
References
-
- Howley P.M. (1991) Role of the human papillomaviruses in human cancer. Cancer Res., 51, 5019–5022. - PubMed
-
- de Villiers E.M. (1994) Human pathogenic papillomavirus types: an update. Curr. Top. Microbiol. Immunol., 186, 1–12. - PubMed
-
- Zur Hausen H. (1996) Papillomavirus infections—a major cause of human cancers. Biochim. Biophys. Acta Rev. Cancer, 1288, 55–78. - PubMed
-
- Zur Hausen H. (2002) Papillomaviruses and cancer: from basic studies to clinical application. Nature Rev. Cancer, 2, 342–350. - PubMed
-
- Munger K. (2002) The role of human papillomaviruses in human cancers. Front. Biosci., 7, 641–649. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

