Hybridization kinetics and thermodynamics of molecular beacons
- PMID: 12582252
- PMCID: PMC150230
- DOI: 10.1093/nar/gkg212
Hybridization kinetics and thermodynamics of molecular beacons
Abstract
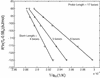
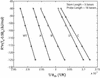
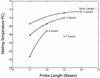
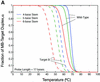
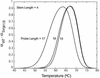
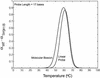
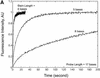
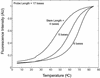
Molecular beacons are increasingly being used in many applications involving nucleic acid detection and quantification. The stem-loop structure of molecular beacons provides a competing reaction for probe-target hybridization that serves to increase probe specificity, which is particularly useful when single-base discrimination is desired. To fully realize the potential of molecular beacons, it is necessary to optimize their structure. Here we report a systematic study of the thermodynamic and kinetic parameters that describe the molecular beacon structure-function relationship. Both probe and stem lengths are shown to have a significant impact on the binding specificity and hybridization kinetic rates of molecular beacons. Specifically, molecular beacons with longer stem lengths have an improved ability to discriminate between targets over a broader range of temperatures. However, this is accompanied by a decrease in the rate of molecular beacon-target hybridization. Molecular beacons with longer probe lengths tend to have lower dissociation constants, increased kinetic rate constants, and decreased specificity. Molecular beacons with very short stems have a lower signal-to-background ratio than molecular beacons with longer stems. These features have significant implications for the design of molecular beacons for various applications.
Figures




Similar articles
-
Structure-function relationships of shared-stem and conventional molecular beacons.Nucleic Acids Res. 2002 Oct 1;30(19):4208-15. doi: 10.1093/nar/gkf536. Nucleic Acids Res. 2002. PMID: 12364599 Free PMC article.
-
Hybridization of 2'-O-methyl and 2'-deoxy molecular beacons to RNA and DNA targets.Nucleic Acids Res. 2003 Mar 15;31(6):5168-74. Nucleic Acids Res. 2003. PMID: 12669716
-
Hybridization of 2'-O-methyl and 2'-deoxy molecular beacons to RNA and DNA targets.Nucleic Acids Res. 2002 Dec 1;30(23):5168-74. Nucleic Acids Res. 2002. Corrected and republished in: Nucleic Acids Res. 2003 Mar 15;31(6):5168-74. PMID: 12466541 Free PMC article. Corrected and republished.
-
DNA probes: applications of the principles of nucleic acid hybridization.Crit Rev Biochem Mol Biol. 1991;26(3-4):227-59. doi: 10.3109/10409239109114069. Crit Rev Biochem Mol Biol. 1991. PMID: 1718662 Review.
-
Molecular beacons: nucleic acid hybridization and emerging applications.J Biomol Struct Dyn. 2001 Dec;19(3):497-504. doi: 10.1080/07391102.2001.10506757. J Biomol Struct Dyn. 2001. PMID: 11790147 Review.
Cited by
-
Dual FRET molecular beacons for mRNA detection in living cells.Nucleic Acids Res. 2004 Apr 14;32(6):e57. doi: 10.1093/nar/gnh062. Nucleic Acids Res. 2004. PMID: 15084672 Free PMC article.
-
Molecular beacons: powerful tools for imaging RNA in living cells.J Nucleic Acids. 2011;2011:741723. doi: 10.4061/2011/741723. Epub 2011 Aug 22. J Nucleic Acids. 2011. PMID: 21876785 Free PMC article.
-
Fluorescence detection of single-nucleotide polymorphisms with a single, self-complementary, triple-stem DNA probe.Angew Chem Int Ed Engl. 2009;48(24):4354-8. doi: 10.1002/anie.200900369. Angew Chem Int Ed Engl. 2009. PMID: 19431180 Free PMC article.
-
Molecular engineering of DNA: molecular beacons.Angew Chem Int Ed Engl. 2009;48(5):856-70. doi: 10.1002/anie.200800370. Angew Chem Int Ed Engl. 2009. PMID: 19065690 Free PMC article. Review.
-
In vitro selection of molecular beacons.Nucleic Acids Res. 2003 Oct 1;31(19):5700-13. doi: 10.1093/nar/gkg764. Nucleic Acids Res. 2003. PMID: 14500834 Free PMC article.
References
-
- Tyagi S. and Kramer,F.R. (1996) Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol., 14, 303–308. - PubMed
-
- Tyagi S., Bratu,D.P. and Kramer,F.R. (1998) Multicolor molecular beacons for allele discrimination. Nat. Biotechnol., 16, 49–53. - PubMed
-
- Kostrikis L.G., Tyagi,S., Mhlanga,M.M., Ho,D.D. and Kramer,F.R. (1998) Spectral genotyping of human alleles. Science, 279, 1228–1229. - PubMed
-
- Piatek A.S., Tyagi,S., Pol,A.C., Telenti,A., Miller,L.P., Kramer,F.R. and Alland,D. (1998) Molecular beacon sequence analysis for detecting drug resistance in Mycobacterium tuberculosis. Nat. Biotechnol., 16, 359–363. - PubMed
-
- Mullah B. and Livak,K. (1999) Efficient automated synthesis of molecular beacons. Nucl. Nucl., 18, 1311–1312.

