Identification and removal of contaminating fluorescence from commercial and in-house printed DNA microarrays
- PMID: 12582263
- PMCID: PMC150250
- DOI: 10.1093/nar/gng018
Identification and removal of contaminating fluorescence from commercial and in-house printed DNA microarrays
Abstract
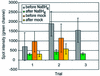
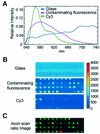
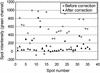
Microarray analysis is a critically important technology for genome-enabled biology, therefore it is essential that the data obtained be reliable. Current software and normalization techniques for microarray analysis rely on the assumption that fluorescent background within spots is essentially the same throughout the glass slide and can be measured by fluorescence surrounding the spots. This assumption is not valid if background fluorescence is spot-localized. Inaccurate estimates of background fluorescence under the spot create a source of error, especially for low expressed genes. We have identified spot-localized, contaminating fluorescence in the Cy3 channel on several commercial and in-house printed microarray slides. We determined through mock hybridizations (without labeled target) that pre-hybridization scans could not be used to predict the contribution of this contaminating fluorescence after hybridization because the change in spot-to-spot fluorescence after hybridization was too variable. Two solutions to this problem were identified. First, allowing 4 h of exposure to air prior to printing on to Corning UltraGAPS slides significantly reduced contaminating fluorescence intensities to approximately the value of the surrounding glass. Alternatively, application of a novel, hyperspectral imaging scanner and multivariate curve resolution algorithms, allowed the spectral contributions of Cy3 signal, glass, and contaminating fluorescence to be distinguished and quantified after hybridization.
Figures






References
-
- Shalon D., Smith,S.J. and Brown,P.O. (1996) A DNA microarray system for analyzing complex DNA samples using two-color fluorescent probe hybridization. Genome Res., 6, 639–645. - PubMed
-
- Schena M., Shalon,D., Davis,R.W. and Brown,P.O. (1995) Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science, 270, 467–470. - PubMed
-
- Quackenbush J. (2001) Computational analysis of microarray data. Nature Rev. Genet., 2, 418–427. - PubMed
-
- Brenner S., Johnson,M., Bridgham,J., Golda,G., Lloyd,D.H., Johnson,D., Luo,S.J., McCurdy,S., Foy,M., Ewan,M. et al. (2000) Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat. Biotechnol., 18, 630–634. - PubMed
-
- Li X.M., Gu,W.K., Mohan,S. and Baylink,D.J. (2002) DNA microarrays: their use and misuse. Microcirculation, 9, 13–22. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

