Endoproteolytic processing of the lymphocytic choriomeningitis virus glycoprotein by the subtilase SKI-1/S1P
- PMID: 12584310
- PMCID: PMC149737
- DOI: 10.1128/jvi.77.5.2866-2872.2003
Endoproteolytic processing of the lymphocytic choriomeningitis virus glycoprotein by the subtilase SKI-1/S1P
Abstract
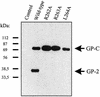
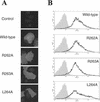
The envelope glycoprotein (GP) of lymphocytic choriomeningitis virus (LCMV) is posttranslationally cleaved into two subunits. We show here that this endoproteolytic processing is not required for transport to the cell surface but is essential for LCMV GP to mediate infectivity of pseudotyped retroviral vectors. By systematic mutational analysis of the LCMV GP cleavage site, we determined that the consensus motif R-(R/K/H)-L-(A/L/S/T/F)(265) is essential for the endoproteolytic processing. In agreement with the identified consensus motif, we show that the cellular subtilase SKI-1/S1P cleaves LCMV GP.
Figures
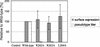


Similar articles
-
The role of proteolytic processing and the stable signal peptide in expression of the Old World arenavirus envelope glycoprotein ectodomain.Virology. 2013 Feb 5;436(1):127-33. doi: 10.1016/j.virol.2012.10.038. Epub 2012 Dec 4. Virology. 2013. PMID: 23218200 Free PMC article.
-
Mechanisms for lymphocytic choriomeningitis virus glycoprotein cleavage, transport, and incorporation into virions.Virology. 2003 Sep 15;314(1):168-78. doi: 10.1016/s0042-6822(03)00421-5. Virology. 2003. PMID: 14517070
-
Oncoretrovirus and lentivirus vectors pseudotyped with lymphocytic choriomeningitis virus glycoprotein: generation, concentration, and broad host range.J Virol. 2002 Feb;76(3):1488-95. doi: 10.1128/jvi.76.3.1488-1495.2002. J Virol. 2002. PMID: 11773421 Free PMC article.
-
The Lassa virus glycoprotein precursor GP-C is proteolytically processed by subtilase SKI-1/S1P.Proc Natl Acad Sci U S A. 2001 Oct 23;98(22):12701-5. doi: 10.1073/pnas.221447598. Epub 2001 Oct 16. Proc Natl Acad Sci U S A. 2001. PMID: 11606739 Free PMC article.
-
[Studies of tissue endoproteases determining viral tropism].Uirusu. 1992 Dec;42(2):119-31. doi: 10.2222/jsv.42.119. Uirusu. 1992. PMID: 1295208 Review. Japanese. No abstract available.
Cited by
-
The role of proteolytic processing and the stable signal peptide in expression of the Old World arenavirus envelope glycoprotein ectodomain.Virology. 2013 Feb 5;436(1):127-33. doi: 10.1016/j.virol.2012.10.038. Epub 2012 Dec 4. Virology. 2013. PMID: 23218200 Free PMC article.
-
Development of Reverse Genetics for the Prototype New World Mammarenavirus Tacaribe Virus.J Virol. 2020 Sep 15;94(19):e01014-20. doi: 10.1128/JVI.01014-20. Print 2020 Sep 15. J Virol. 2020. PMID: 32669332 Free PMC article.
-
Endocytosis plays a critical role in proteolytic processing of the Hendra virus fusion protein.J Virol. 2005 Oct;79(20):12643-9. doi: 10.1128/JVI.79.20.12643-12649.2005. J Virol. 2005. PMID: 16188966 Free PMC article.
-
Characterization of Lassa virus glycoprotein oligomerization and influence of cholesterol on virus replication.J Virol. 2010 Jan;84(2):983-92. doi: 10.1128/JVI.02039-09. Epub 2009 Nov 4. J Virol. 2010. PMID: 19889753 Free PMC article.
-
Comparative analysis of disease pathogenesis and molecular mechanisms of New World and Old World arenavirus infections.J Gen Virol. 2014 Jan;95(Pt 1):1-15. doi: 10.1099/vir.0.057000-0. Epub 2013 Sep 25. J Gen Virol. 2014. PMID: 24068704 Free PMC article. Review.
References
-
- Asper, M., P. Hofmann, C. Osmann, J. Funk, C. Metzger, M. Bruns, F. J. Kaup, H. Schmitz, and S. Gunther. 2001. First outbreak of callitrichid hepatitis in Germany: genetic characterization of the causative lymphocytic choriomeningitis virus strains. Virology 284:203-213. - PubMed
-
- Auperin, D. D., D. R. Sasso, and J. B. McCormick. 1986. Nucleotide sequence of the glycoprotein gene and intergenic region of the Lassa virus S genome RNA. Virology 154:155-167. - PubMed
-
- Basak, A., M. Chretien, and N. G. Seidah. 2002. A rapid fluorometric assay for the proteolytic activity of SKI-1/S1P based on the surface glycoprotein of the hemorrhagic fever Lassa virus. FEBS Lett. 514:333-339. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

