Efficient replication of hepatitis C virus genotype 1a RNAs in cell culture
- PMID: 12584342
- PMCID: PMC149761
- DOI: 10.1128/jvi.77.5.3181-3190.2003
Efficient replication of hepatitis C virus genotype 1a RNAs in cell culture
Abstract
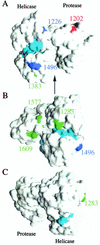
Hepatitis C virus (HCV) genotype 1 (subtypes 1a and 1b) is responsible for the majority of treatment-resistant liver disease worldwide. Thus far, efficient HCV RNA replication has been observed only for subgenomic and full-length RNAs derived from genotype 1b isolates. Here, we report the establishment of efficient RNA replication systems for genotype 1a strain H77. Replication of subgenomic and full-length H77 1a RNAs required the highly permissive Huh-7.5 hepatoma subline and adaptive amino acid substitutions in both NS3 and NS5A. Replication could be detected by RNA quantification, fluorescence-activated cell sorting, and metabolic labeling of HCV-specific proteins. Replication efficiencies were similar for subgenomic and full-length RNAs and were most efficient for HCV RNAs lacking heterologous RNA elements. Interestingly, both subtype 1a and 1b NS3 adaptive mutations are surface exposed and present on only one face of the NS3 structure. The cell culture-adapted subtype 1a replicons should be useful for basic replication studies and for antiviral development. These results are also encouraging for the development of adapted replicons for the remaining HCV genotypes.
Figures






References
-
- Blight, K. J., A. Grakoui, H. L. Hanson, and C. M. Rice. 2002. The molecular biology of hepatitis C virus, p. 81-108. In J.-H. J. Ou (ed.), Hepatitis viruses. Kluwer Academic Publishers, Boston, Mass.
-
- Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. - PubMed
-
- Cornberg, M., H. Wedemeyer, and M. P. Manns. 2001. Hepatitis C: therapeutic perspectives. Forum 11:154-162. - PubMed
-
- Cornberg, M., H. Wedemeyer, and M. P. Manns. 2002. Treatment of chronic hepatitis C with PEGylated interferon and ribavirin. Curr. Gastroenterol. Rep. 4:23-30. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

