In exponentially growing Saccharomyces cerevisiae cells, rRNA synthesis is determined by the summed RNA polymerase I loading rate rather than by the number of active genes
- PMID: 12588976
- PMCID: PMC151703
- DOI: 10.1128/MCB.23.5.1558-1568.2003
In exponentially growing Saccharomyces cerevisiae cells, rRNA synthesis is determined by the summed RNA polymerase I loading rate rather than by the number of active genes
Abstract
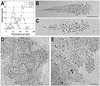
Genes encoding rRNA are multicopy and thus could be regulated by changing the number of active genes or by changing the transcription rate per gene. We tested the hypothesis that the number of open genes is limiting rRNA synthesis by using an electron microscopy method that allows direct counting of the number of active genes per nucleolus and the number of polymerases per active gene. Two strains of Saccharomyces cerevisiae were analyzed during exponential growth: a control strain with a typical number of rRNA genes ( approximately 143 in this case) and a strain in which the rRNA gene number was reduced to approximately 42 but which grows as well as controls. In control strains, somewhat more than half of the genes were active and the mean number of polymerases/gene was approximately 50 +/- 20. In the 42-copy strain, all rRNA genes were active with a mean number of 100 +/- 29 polymerases/gene. Thus, an equivalent number of polymerases was active per nucleolus in the two strains, though the number of active genes varied by twofold, showing that overall initiation rate, and not the number of active genes, determines rRNA transcription rate during exponential growth in yeast. Results also allow an estimate of elongation rate of approximately 60 nucleotides/s for yeast Pol I and a reinitiation rate of less than 1 s on the most heavily transcribed genes.
Figures




References
-
- Cavanaugh, A. H., W. M. Hempel, L. J. Taylor, V. Rogalsky, G. Todorov, and L. I. Rothblum. 1995. Activity of RNA polymerase I transcription factor UBF blocked by Rb gene product. Nature 374:177-180. - PubMed
-
- Cavanaugh, A. H., I. Hirschler-Laszkiewicz, Q. Hu, M. Dundr, T. Smink, T. Misteli, and L. I. Rothblum. 2002. Rrn3 phosphorylation is a regulatory checkpoint for ribosome biogenesis. J. Biol. Chem. 277:27423-27432. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
