Nob1p is required for cleavage of the 3' end of 18S rRNA
- PMID: 12588997
- PMCID: PMC151717
- DOI: 10.1128/MCB.23.5.1798-1807.2003
Nob1p is required for cleavage of the 3' end of 18S rRNA
Abstract
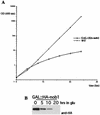
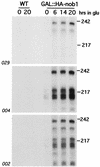
We report the characterization of a novel factor, Nob1p (Yor056c), which is essential for the synthesis of 40S ribosome subunits. Genetic depletion of Nob1p strongly inhibits the processing of the 20S pre-rRNA to the mature 18S rRNA, leading to the accumulation of high levels of the 20S pre-rRNA together with novel degradation intermediates. 20S processing occurs within a pre-40S particle after its export from the nucleus to the cytoplasm. Consistent with a direct role in this cleavage, Nob1p was shown to be associated with the pre-40S particle and to be present in both the nucleus and the cytoplasm. This suggests that Nob1p accompanies the pre-40S ribosomes during nuclear export. Pre-40S export is not, however, inhibited by depletion of Nob1p.
Figures


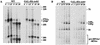


Similar articles
-
PIN domain of Nob1p is required for D-site cleavage in 20S pre-rRNA.RNA. 2004 Nov;10(11):1698-701. doi: 10.1261/rna.7123504. Epub 2004 Sep 23. RNA. 2004. PMID: 15388878 Free PMC article.
-
The novel ATP-binding cassette protein ARB1 is a shuttling factor that stimulates 40S and 60S ribosome biogenesis.Mol Cell Biol. 2005 Nov;25(22):9859-73. doi: 10.1128/MCB.25.22.9859-9873.2005. Mol Cell Biol. 2005. PMID: 16260602 Free PMC article.
-
Yeast Krr1p physically and functionally interacts with a novel essential Kri1p, and both proteins are required for 40S ribosome biogenesis in the nucleolus.Mol Cell Biol. 2000 Nov;20(21):7971-9. doi: 10.1128/MCB.20.21.7971-7979.2000. Mol Cell Biol. 2000. PMID: 11027267 Free PMC article.
-
The Saccharomyces cerevisiae TIF6 gene encoding translation initiation factor 6 is required for 60S ribosomal subunit biogenesis.Mol Cell Biol. 2001 Mar;21(5):1453-62. doi: 10.1128/MCB.21.5.1453-1462.2001. Mol Cell Biol. 2001. PMID: 11238882 Free PMC article.
-
Nuclear export and cytoplasmic maturation of ribosomal subunits.FEBS Lett. 2007 Jun 19;581(15):2783-93. doi: 10.1016/j.febslet.2007.05.013. Epub 2007 May 11. FEBS Lett. 2007. PMID: 17509569 Review.
Cited by
-
atBRX1-1 and atBRX1-2 are involved in an alternative rRNA processing pathway in Arabidopsis thaliana.RNA. 2015 Mar;21(3):415-25. doi: 10.1261/rna.047563.114. Epub 2015 Jan 20. RNA. 2015. PMID: 25605960 Free PMC article.
-
Domain definition and interaction mapping for the endonuclease complex hNob1/hPno1.RNA Biol. 2018;15(9):1174-1180. doi: 10.1080/15476286.2018.1517013. Epub 2018 Sep 18. RNA Biol. 2018. PMID: 30176151 Free PMC article.
-
[Eukaryotic translation elongation factor 1A1 positively regulates NOB1 expression to promote invasion and metastasis of hepatocellular carcinoma cells in vitro].Nan Fang Yi Ke Da Xue Xue Bao. 2018 Sep 30;38(10):1195-1202. doi: 10.3969/j.issn.1673-4254.2018.10.07. Nan Fang Yi Ke Da Xue Xue Bao. 2018. PMID: 30377124 Free PMC article. Chinese.
-
Impact of two neighbouring ribosomal protein clusters on biogenesis factor binding and assembly of yeast late small ribosomal subunit precursors.PLoS One. 2019 Jan 17;14(1):e0203415. doi: 10.1371/journal.pone.0203415. eCollection 2019. PLoS One. 2019. PMID: 30653518 Free PMC article.
-
Mrd1p binds to pre-rRNA early during transcription independent of U3 snoRNA and is required for compaction of the pre-rRNA into small subunit processomes.Nucleic Acids Res. 2008 Aug;36(13):4364-80. doi: 10.1093/nar/gkn384. Epub 2008 Jun 27. Nucleic Acids Res. 2008. PMID: 18586827 Free PMC article.
References
-
- Amberg, D. C., A. L. Goldstein, and C. N. Cole. 1992. Isolation and characterization of RAT1: an essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes Dev. 6:1173-1189. - PubMed
-
- Baßler, J., P. Grandi, O. Gadal, T. Leßmann, E. Petfalski, D. Tollervey, J. Lechner, and E. C. Hurt. 2001. Identification of a pre-ribosomal particle that is closely linked to nuclear export. Mol. Cell 8:517-529. - PubMed
-
- Brand, R. C., J. Klootwijk, T. J. M. van Steanbergen, A. J. de Kok, and R. J. Planta. 1977. Secondary methylation of yeast ribosomal precursor RNA. Eur. J. Biochem. 75:311-318. - PubMed
-
- Clissold, P. M., and C. P. Ponting. 2000. PIN domains in nonsense-mediated mRNA decay and RNAi. Curr. Biol. 10:R888-R890. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
