Compensatory mutations cause excess of antagonistic epistasis in RNA secondary structure folding
- PMID: 12590655
- PMCID: PMC149451
- DOI: 10.1186/1471-2148-3-3
Compensatory mutations cause excess of antagonistic epistasis in RNA secondary structure folding
Abstract
Background: The rate at which fitness declines as an organism's genome accumulates random mutations is an important variable in several evolutionary theories. At an intuitive level, it might seem natural that random mutations should tend to interact synergistically, such that the rate of mean fitness decline accelerates as the number of random mutations is increased. However, in a number of recent studies, a prevalence of antagonistic epistasis (the tendency of multiple mutations to have a mitigating rather than reinforcing effect) has been observed.
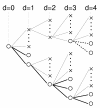
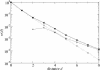
Results: We studied in silico the net amount and form of epistatic interactions in RNA secondary structure folding by measuring the fraction of neutral mutants as a function of mutational distance d. We found a clear prevalence of antagonistic epistasis in RNA secondary structure folding. By relating the fraction of neutral mutants at distance d to the average neutrality at distance d, we showed that this prevalence derives from the existence of many compensatory mutations at larger mutational distances.
Conclusions: Our findings imply that the average direction of epistasis in simple fitness landscapes is directly related to the density with which fitness peaks are distributed in these landscapes.
Figures





References
-
- Phillips P, Otto S, Whitlock M. Beyond the average: the evolutionary importance of gene interactions and variability of epistatic effects. In: Wolf J, Brodie E, Wade M, editor. Epistasis and the Evolutionary Process. Oxford, UK, Oxford University Press; 2000. pp. 20–38.
-
- Whitlock MC, Phillips PC, Moore FB-G, Tonsor SJ. Multiple fitness peaks and epistasis. Annu Rev Ecol Syst. 1995;26:601–629. doi: 10.1146/annurev.es.26.110195.003125. - DOI
-
- Wolf J, Brodie E, Wade M, editors Epistasis and the Evolutionary Process. Oxford, UK, Oxford University Press. 2000.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

