Transgenic expression of the activating natural killer receptor Ly49H confers resistance to cytomegalovirus in genetically susceptible mice
- PMID: 12591908
- PMCID: PMC2193863
- DOI: 10.1084/jem.20021713
Transgenic expression of the activating natural killer receptor Ly49H confers resistance to cytomegalovirus in genetically susceptible mice
Abstract
Natural resistance to infection with mouse cytomegalovirus (MCMV) is controlled by a dominant locus, Cmv1. Cmv1 is linked to the Ly49 family of natural killer receptors on distal chromosome 6. While some studies localized Cmv1 as distal to the Ly49 gene cluster, genetic and functional analysis identified Ly49h as a pivotal factor in resistance to MCMV. The role of these two independent genomic domains in MCMV resistance was evaluated by functional complementation using transgenesis of bacterial artificial chromosomes (BAC) in genetically susceptible mice. Phenotypic and genetic characterization of the transgenic animals traced the resistance gene to a single region spanning the Ly49h gene. The appearance of the Ly49H protein in NK cells of transgenic mice coincided with the emergence of MCMV resistance, and there was a threshold Ly49H protein level associated with full recovery. Finally, transgenic expression of Ly49H in the context of either of the two independent susceptibility alleles, Cmv1(sBALB) or Cmv1(sFVB), conferred resistance to MCMV infection. These results demonstrate that Ly49h is necessary and sufficient to confer MCMV resistance, and formally demonstrate allelism between Cmv1 and Ly49h. This panel of transgenic animals provides a unique resource to study possible pleiotropic effect of Cmv1.
Figures



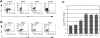

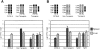

References
-
- Chalmer, J.E., J.S. Mackenzie, and N.F. Stanley. 1977. Resistance to murine cytomegalovirus linked to the major histocompatibility complex of the mouse. J. Gen. Virol. 37:107–114. - PubMed
-
- Allan, J.E., and G.R. Shellam. 1984. Genetic control of murine cytomegalovirus infection: virus titres in resistant and susceptible strains of mice. Arch. Virol. 81:139–150. - PubMed
-
- Scalzo, A.A., N.A. Fitzgerald, C.R. Wallace, A.E. Gibbons, Y.C. Smart, R.C. Burton, and G.R. Shellam. 1992. The effect of the Cmv-1 resistance gene, which is linked to the natural killer cell gene complex, is mediated by natural killer cells. J. Immunol. 149:581–589. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

