The molecular function of Ase1p: evidence for a MAP-dependent midzone-specific spindle matrix. Microtubule-associated proteins
- PMID: 12591913
- PMCID: PMC2173742
- DOI: 10.1083/jcb.200210021
The molecular function of Ase1p: evidence for a MAP-dependent midzone-specific spindle matrix. Microtubule-associated proteins
Abstract
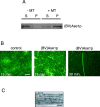
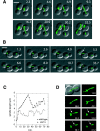
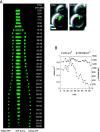
The midzone is the domain of the mitotic spindle that maintains spindle bipolarity during anaphase and generates forces required for spindle elongation (anaphase B). Although there is a clear role for microtubule (MT) motor proteins at the spindle midzone, less is known about how microtubule-associated proteins (MAPs) contribute to midzone organization and function. Here, we report that budding yeast Ase1p is a member of a conserved family of midzone-specific MAPs. By size exclusion chromatography and velocity sedimentation, both Ase1p in extracts and purified Ase1p behaved as a homodimer. Ase1p bound and bundled MTs in vitro. By live cell microscopy, loss of Ase1p resulted in a specific defect: premature spindle disassembly in mid-anaphase. Furthermore, when overexpressed, Ase1p was sufficient to trigger spindle elongation in S phase-arrested cells. FRAP revealed that Ase1p has both a very slow rate of turnover within the midzone and limited lateral diffusion along spindle MTs. We propose that Ase1p functions as an MT cross-bridge that imparts matrix-like characteristics to the midzone. MT-dependent networks of spindle midzone MAPs may be one molecular basis for the postulated spindle matrix.
Figures





References
-
- Adams, R.R., M. Carmena, and W.C. Earnshaw. 2001. Chromosomal passengers and the (aurora) ABCs of mitosis. Trends Cell Biol. 11:49–54. - PubMed
-
- Aist, J.R., C.J. Bayles, W. Tao, and M.W. Berns. 1991. Direct experimental evidence for the existence, structural basis, and function of astral forces during anaphase B in vivo. J. Cell Sci. 100:279–288. - PubMed
-
- Aist, J.R., H. Liang, and M.W. Berns. 1993. Astral and spindle forces in PtK2 cells during anaphase B: a laser microbeam study. J. Cell Sci. 104:1207–1216. - PubMed
-
- Altschul, S.F., and W. Gish. 1996. Local alignment statistics. Methods Enzymol. 266:460–480. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

