The inner membrane protein Mdm33 controls mitochondrial morphology in yeast
- PMID: 12591915
- PMCID: PMC2173741
- DOI: 10.1083/jcb.200211113
The inner membrane protein Mdm33 controls mitochondrial morphology in yeast
Abstract
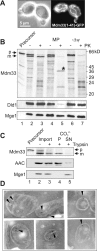
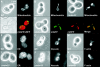
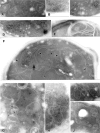
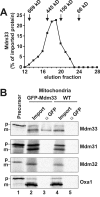
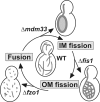
Mitochondrial distribution and morphology depend on MDM33, a Saccharomyces cerevisiae gene encoding a novel protein of the mitochondrial inner membrane. Cells lacking Mdm33 contain ring-shaped, mostly interconnected mitochondria, which are able to form large hollow spheres. On the ultrastructural level, these aberrant organelles display extremely elongated stretches of outer and inner membranes enclosing a very narrow matrix space. Dilated parts of Delta mdm33 mitochondria contain well-developed cristae. Overexpression of Mdm33 leads to growth arrest, aggregation of mitochondria, and generation of aberrant inner membrane structures, including septa, inner membrane fragments, and loss of inner membrane cristae. The MDM33 gene is required for the formation of net-like mitochondria in mutants lacking components of the outer membrane fission machinery, and mitochondrial fusion is required for the formation of extended ring-like mitochondria in cells lacking the MDM33 gene. The Mdm33 protein assembles into an oligomeric complex in the inner membrane where it performs homotypic protein-protein interactions. Our results indicate that Mdm33 plays a distinct role in the mitochondrial inner membrane to control mitochondrial morphology. We propose that Mdm33 is involved in fission of the mitochondrial inner membrane.
Figures



Similar articles
-
Interaction of MDM33 with mitochondrial inner membrane homeostasis pathways in yeast.Sci Rep. 2015 Dec 16;5:18344. doi: 10.1038/srep18344. Sci Rep. 2015. PMID: 26669658 Free PMC article.
-
The intramitochondrial dynamin-related GTPase, Mgm1p, is a component of a protein complex that mediates mitochondrial fusion.J Cell Biol. 2003 Feb 3;160(3):303-11. doi: 10.1083/jcb.200209015. J Cell Biol. 2003. PMID: 12566426 Free PMC article.
-
Mgm1p, a dynamin-related GTPase, is essential for fusion of the mitochondrial outer membrane.Mol Biol Cell. 2003 Jun;14(6):2342-56. doi: 10.1091/mbc.e02-12-0788. Epub 2003 Feb 6. Mol Biol Cell. 2003. PMID: 12808034 Free PMC article.
-
The enigmatic role of Mim1 in mitochondrial biogenesis.Eur J Cell Biol. 2010 Feb-Mar;89(2-3):212-5. doi: 10.1016/j.ejcb.2009.11.002. Epub 2009 Nov 26. Eur J Cell Biol. 2010. PMID: 19944477 Review.
-
Protein insertion into the inner membrane of mitochondria.IUBMB Life. 2003 Apr-May;55(4-5):219-25. doi: 10.1080/1521654031000123349. IUBMB Life. 2003. PMID: 12880202 Review.
Cited by
-
Intersection between mitochondrial permeability pores and mitochondrial fusion/fission.Neurochem Res. 2007 Apr-May;32(4-5):917-29. doi: 10.1007/s11064-006-9252-2. Epub 2007 Mar 7. Neurochem Res. 2007. PMID: 17342412 Review.
-
Phospholipid ebb and flow makes mitochondria go.J Cell Biol. 2020 Aug 3;219(8):e202003131. doi: 10.1083/jcb.202003131. J Cell Biol. 2020. PMID: 32614384 Free PMC article. Review.
-
Mitochondrial dynamics in the adult cardiomyocytes: which roles for a highly specialized cell?Front Physiol. 2013 May 10;4:102. doi: 10.3389/fphys.2013.00102. eCollection 2013. Front Physiol. 2013. PMID: 23675354 Free PMC article.
-
A mitochondrial-focused genetic interaction map reveals a scaffold-like complex required for inner membrane organization in mitochondria.J Cell Biol. 2011 Oct 17;195(2):323-40. doi: 10.1083/jcb.201107053. Epub 2011 Oct 10. J Cell Biol. 2011. PMID: 21987634 Free PMC article.
-
Electron microscopic analysis of a spherical mitochondrial structure.J Biol Chem. 2012 Dec 7;287(50):42373-8. doi: 10.1074/jbc.M112.413674. Epub 2012 Oct 23. J Biol Chem. 2012. PMID: 23093403 Free PMC article.
References
-
- Andaluz, E., J.J. Coque, R. Cueva, and G. Larriba. 2001. Sequencing of a 4.3 kbp region of chromosome 2 of Candida albicans reveals the presence of homologues of SHE9 from Saccharomyces cerevisiae and of bacterial phosphatidylinositol-phospholipase C. Yeast. 18:711–721. - PubMed
-
- Bereiter-Hahn, J. 1990. Behavior of mitochondria in the living cell. Int. Rev. Cytol. 122:1–63. - PubMed
-
- Boldogh, I.R., H.-C. Yang, and L.A. Pon. 2001. Mitochondrial inheritance in budding yeast. Traffic. 2:368–374. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

