Structure of a cholesterol-binding protein deficient in Niemann-Pick type C2 disease
- PMID: 12591954
- PMCID: PMC151372
- DOI: 10.1073/pnas.0437840100
Structure of a cholesterol-binding protein deficient in Niemann-Pick type C2 disease
Abstract
Niemann-Pick disease type C2 (NP-C2) is a fatal hereditary disease characterized by accumulation of low-density lipoprotein-derived cholesterol in lysosomes. Here we report the 1.7-A resolution crystal structure of the cholesterol-binding protein deficient in this disease, NPC2, and the characterization of its ligand binding properties. Human NPC2 binds the cholesterol analog dehydroergosterol with submicromolar affinity at both acidic and neutral pH. NPC2 has an Ig-like fold stabilized by three disulfide bonds. The structure of the bovine protein reveals a loosely packed region penetrating from the surface into the hydrophobic core that forms adjacent small cavities with a total volume of approximately 160 A(3). We propose that this region represents the incipient cholesterol-binding site that dilates to accommodate an approximately 740-A(3) cholesterol molecule.
Figures
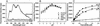

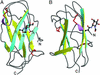
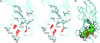

References
-
- Goldstein J L, Hobbs H H, Brown M S. In: The Metabolic and Molecular Bases of Inherited Disease. Scriver C R, Beaudet A L, Sly W S, Valle D, editors. III. New York: McGraw–Hill; 2001. pp. 2863–2913.
-
- Patterson M C, Vanier M T, Suzuki K, Morris J A, Carstea E, Neufeld E B, Blanchette-Mackie J E, Pentchev P G. In: The Metabolic and Molecular Bases of Inherited Disease. Scriver C R, Beaudet A L, Sly W S, Valle D, editors. III. New York: McGraw–Hill; 2001. pp. 3611–3633.
-
- Carstea E D, Morris J A, Coleman K G, Loftus S K, Zhang D, Cummings C, Gu J, Rosenfeld M A, Pavan W J, Krizman D B, et al. Science. 1997;277:228–231. - PubMed
-
- Ioannou Y A. Nat Rev Mol Cell Biol. 2001;2:657–668. - PubMed
-
- Davies J P, Chen F W, Ioannou Y A. Science. 2000;290:2295–2298. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

