Recurring features of local tertiary structural elements in RNA molecules exemplified by hepatitis D virus RNA
- PMID: 12592001
- PMCID: PMC1370394
- DOI: 10.1261/rna.2173903
Recurring features of local tertiary structural elements in RNA molecules exemplified by hepatitis D virus RNA
Abstract
Elements of local tertiary structure in RNA molecules are important in understanding structure-function relationships. The loop E motif, first identified in several eukaryotic RNAs at functional sites which share an exceptional propensity for UV crosslinking between specific bases, was subsequently shown to have a characteristic tertiary structure. Common sequences and secondary structures have allowed other examples of the E-loop motif to be recognized in a number of RNAs at sites of protein binding or other biological function. We would like to know if more elements of local tertiary structure, in addition to the E-loop, can be identified by such common features. The highly structured circular RNA genome of the hepatitis D virus (HDV) provides an ideal test molecule because it has extensive internal structure, a UV-crosslinkable tertiary element, and specific sites for functional interactions with proteins including host PKR. We have now found a UV-crosslinkable element of local tertiary structure in antigenomic HDV RNA which, although differing from the E-loop, has a very similar pattern of sequence and secondary structure to the UV-crosslinkable element found in the genomic strand. Despite the fact that the two structures map close to one another, the sequences comprising them are not the templates for each other. Instead, the template regions for each element are additional sites for potential higher order structure on their respective complementary strands. This wealth of recurring sequences interspersed with base-paired stems provides a context to examine other RNA species for such features and their correlations with biological function.
Figures

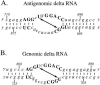

References
-
- Allison, L.A., Romaniuk, P.J., and Bakken, A.H. 1991. RNA–protein interactions of stored 5S RNA with TFIIIA and ribosomal protein L5 during Xenopus oogenesis. Dev. Biol. 144: 129–144. - PubMed
-
- Barrell, B.J. 1971. Fractionation and sequence analysis of radioactive nucleotides. In Procedures in nucleic acids research (eds. G.L. Cantoni and D.R. Davies), Vol. 2, pp. 751–779. Harper & Row, New York.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
