Two reactions of Haloferax volcanii RNA splicing enzymes: joining of exons and circularization of introns
- PMID: 12592006
- PMCID: PMC1370399
- DOI: 10.1261/rna.2118203
Two reactions of Haloferax volcanii RNA splicing enzymes: joining of exons and circularization of introns
Abstract
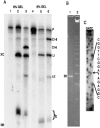
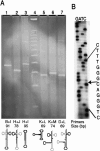
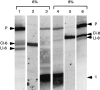
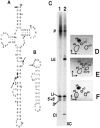
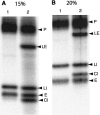
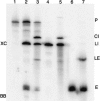
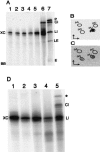
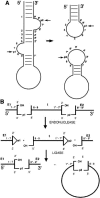
Archaeal RNA splicing involves at least two protein enzymes, a specific endonuclease and a specific ligase. The endonuclease recognizes and cleaves within a characteristic bulge-helix-bulge (BHB) structure formed by pairing of the regions near the two exon-intron junctions, producing 2',3'-cyclic phosphate and 5'-hydroxyl termini. The ligase joins the exons and converts the cyclic phosphate into junction phosphate. The ligated product contains a seven-base hairpin loop, in which the splice junction is in between the two 3' terminal residues of the loop. Archaeal splicing endonucleases are also involved in rRNA processing, cutting within the BHB structures formed by pairing of the 5' and 3' flanking regions of the rRNAs. Large free introns derived from pre-rRNAs have been observed as stable and abundant circular RNAs in certain Crenarchaeota, a kingdom in the domain Archaea. In the present study, we show that the cells of Haloferax volcanii, a Euryarchaeote, contain circular RNAs formed by 3',5'-phosphodiester linkage between the two termini of the introns derived from their pre-tRNAs. H. volcanii ligase, in vitro, can also circularize both endonuclease-cleaved introns, and non-endonuclease-produced substrates. Exon joining and intron circularization are mechanistically similar ligation reactions that can occur independently. The size of the ligated hairpin loop and position of the splice junction within this loop can be changed in in vitro ligation reactions. Overall, archaeal RNA splicing seems to involve two sets of two symmetric transesterification reactions each.
Figures
References
-
- Abelson, J., Trotta, C.R., and Li, H. 1998. tRNA splicing. J. Biol. Chem. 273: 12685–12688. - PubMed
-
- Arn, E.A. and Abelson, J. 1998. RNA ligases: Function, mechanism, and sequence conservation. In RNA structure and function (eds. R.W. Simons and M. Grunberg-Manago), pp. 695–726. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
-
- Dalgaard, J.Z. and Garrett, R.A. 1992. Protein-coding introns from the 23S rRNA-encoding gene from stable circles in the hyperthermophilic archaeon Pyrobaculum organotrophum. Gene 121: 103–110. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
