RNA interference of influenza virus production by directly targeting mRNA for degradation and indirectly inhibiting all viral RNA transcription
- PMID: 12594334
- PMCID: PMC151407
- DOI: 10.1073/pnas.0437841100
RNA interference of influenza virus production by directly targeting mRNA for degradation and indirectly inhibiting all viral RNA transcription
Abstract
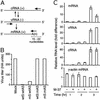
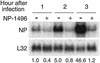
Influenza A virus causes widespread infection in the human respiratory tract, but existing vaccines and drug therapy are of limited value. Here we show that short interfering RNAs (siRNAs) specific for conserved regions of the viral genome can potently inhibit influenza virus production in both cell lines and embryonated chicken eggs. The inhibition depends on the presence of a functional antisense strand in the siRNA duplex, suggesting that viral mRNA is the target of RNA interference. However, siRNA specific for nucleocapsid (NP) or a component of the RNA transcriptase (PA) abolished the accumulation of not only the corresponding mRNA but also virion RNA and its complementary RNA. These siRNAs also broadly inhibited the accumulation of other viral, but not cellular, RNAs. The findings reveal that newly synthesized NP and PA proteins are required for influenza virus transcription and replication and provide a basis for the development of siRNAs as prophylaxis and therapy for influenza infection in humans.
Figures


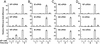
References
-
- Lamb R A, Krug R M. In: Fundamental Virology. Knipe D M, Howley P M, editors. Philadelphia: Lippincott Williams & Wilkins; 2001. pp. 725–770.
-
- Fukuda K, Bridges C B, Brammer T L, Izurieta H S, Cox N J. Morbid Mortal Wkly Rep. 1999;48:1–28.
-
- Patterson K D, Pyle G F. Bull Hist Med. 1991;65:4–21. - PubMed
-
- Webster R G, Laver W G, Air G M, Schild G C. Nature. 1982;296:115–121. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- AI44477/AI/NIAID NIH HHS/United States
- AI40146/AI/NIAID NIH HHS/United States
- CA42063/CA/NCI NIH HHS/United States
- R01 CA060686/CA/NCI NIH HHS/United States
- CA60686/CA/NCI NIH HHS/United States
- P01 CA042063/CA/NCI NIH HHS/United States
- AI32486/AI/NIAID NIH HHS/United States
- R37 GM034277/GM/NIGMS NIH HHS/United States
- R01 GM034277/GM/NIGMS NIH HHS/United States
- GM34277/GM/NIGMS NIH HHS/United States
- AI50631/AI/NIAID NIH HHS/United States
- AI44478/AI/NIAID NIH HHS/United States
- R01 AI050631/AI/NIAID NIH HHS/United States
- R01 AI040146/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

