A triple-mutated allele of granzyme B incapable of inducing apoptosis
- PMID: 12594335
- PMCID: PMC151380
- DOI: 10.1073/pnas.0437935100
A triple-mutated allele of granzyme B incapable of inducing apoptosis
Abstract
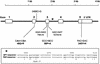
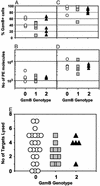
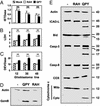
Granzyme B (GzmB) is a serine protease involved in many pathologies, including viral infections, autoimmunity, transplant rejection, and antitumor immunity. To measure the extent of genetic variation in GzmB, we screened the GzmB gene for polymorphisms and defined a frequently represented triple-mutated GzmB allele. In this variant, three amino acids of the mature protein Q(48)P(88)Y(245) are mutated to R(48)A(88)H(245). In CD8(+) cytotoxic T lymphocytes, GzmB was expressed at similar levels in QPY homozygous, QPY/RAH heterozygous, and RAH homozygous individuals, demonstrating that RAH GzmB is a stable protein. Active RAH GzmB expressed in glioblastoma cell lines displayed proteolytic activity, but in contrast to QPY GzmB, it did not accumulate in the nucleus and was unable to induce Bid cleavage, cytochrome c release, or apoptosis. Molecular modeling showed that the three amino acid substitutions clustered near the C-terminal alpha-helix of the protein, indicating that this region of the protein may be involved in the intracellular targeting of GzmB. The triple-mutated GzmB allele that we describe appears to be incapable of inducing apoptosis in tumor cell lines, and its presence could, therefore, influence both the prognosis of cancer patients and the success rates of antitumor cellular immunotherapy.
Figures


References
-
- Peitsch M C, Tschopp J. Methods Enzymol. 1994;244:80–87. - PubMed
-
- Berthou C, Michel L, Soulie A, Jean-Louis F, Flageul B, Dubertret L, Sigaux F, Zhang Y, Sasportes M. J Immunol. 1997;159:5293–5300. - PubMed
-
- Masson D, Tschopp J. Cell. 1987;49:679–685. - PubMed
-
- Motyka B, Korbutt G, Pinkoski M J, Heibein J A, Caputo A, Hobman M, Barry M, Shostak I, Sawchuk T, Holmes C F, et al. Cell. 2000;103:491–500. - PubMed
-
- Pinkoski M J, Hobman M, Heibein J A, Tomaselli K, Li F, Seth P, Froelich C J, Bleackley R C. Blood. 1998;92:1044–1054. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

