Mapping the triphosphatase active site of baculovirus mRNA capping enzyme LEF4 and evidence for a two-metal mechanism
- PMID: 12595553
- PMCID: PMC149837
- DOI: 10.1093/nar/gkg244
Mapping the triphosphatase active site of baculovirus mRNA capping enzyme LEF4 and evidence for a two-metal mechanism
Abstract
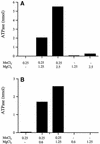
The 464-amino acid baculovirus LEF4 protein is a bifunctional mRNA capping enzyme with triphosphatase and guanylyltransferase activities. The N-terminal half of LEF4 constitutes an autonomous triphosphatase catalytic domain. The LEF4 triphosphatase belongs to a family of metal-dependent phosphohydrolases, which includes the RNA triphosphatases of fungi, protozoa, Chlorella virus and poxviruses. The family is defined by two glutamate-containing motifs (A and C), which form a metal-binding site. Most of the family members resemble the fungal and Chlorella virus enzymes, which have a complex active site located within the hydrophilic interior of a topologically closed eight stranded beta barrel (the so-called 'triphosphate tunnel'). Here we probed whether baculovirus LEF4 is a member of the tunnel subfamily, via mutational mapping of amino acids required for triphosphatase activity. We identified four new essential side chains in LEF4 via alanine scanning and illuminated structure-activity relationships by conservative substitutions. Our results, together with previous mutational data, highlight five acidic and four basic amino acids that are likely to comprise the LEF4 triphosphatase active site (Glu9, Glu11, Arg51, Arg53, Glu97, Lys126, Arg179, Glu181 and Glu183). These nine essential residues are conserved in LEF4 orthologs from all strains of baculoviruses. We discerned no pattern of clustering of the catalytic residues of the baculovirus triphosphatase that would suggest structural similarity to the tunnel proteins (exclusive of motifs A and C). However, there is similarity to the active site of vaccinia RNA triphosphatase. We infer that the baculovirus and poxvirus triphosphatases are a distinct lineage within the metal-dependent RNA triphosphatase family. Synergistic activation of the LEF4 triphosphatase by manganese and magnesium suggests a two-metal mechanism of gamma phosphate hydrolysis.
Figures



Similar articles
-
Mapping the active site of vaccinia virus RNA triphosphatase.Virology. 2003 Apr 25;309(1):125-34. doi: 10.1016/s0042-6822(03)00002-3. Virology. 2003. PMID: 12726733
-
Mutational analysis of baculovirus capping enzyme Lef4 delineates an autonomous triphosphatase domain and structural determinants of divalent cation specificity.J Biol Chem. 2001 Dec 7;276(49):45522-9. doi: 10.1074/jbc.M107615200. Epub 2001 Sep 11. J Biol Chem. 2001. PMID: 11553638
-
Yeast and viral RNA 5' triphosphatases comprise a new nucleoside triphosphatase family.J Biol Chem. 1998 Dec 18;273(51):34151-6. doi: 10.1074/jbc.273.51.34151. J Biol Chem. 1998. PMID: 9852075
-
RNA capping enzyme and DNA ligase: a superfamily of covalent nucleotidyl transferases.Mol Microbiol. 1995 Aug;17(3):405-10. doi: 10.1111/j.1365-2958.1995.mmi_17030405.x. Mol Microbiol. 1995. PMID: 8559059 Review.
-
The mRNA capping apparatus as drug target and guide to eukaryotic phylogeny.Cold Spring Harb Symp Quant Biol. 2001;66:301-12. doi: 10.1101/sqb.2001.66.301. Cold Spring Harb Symp Quant Biol. 2001. PMID: 12762032 Review. No abstract available.
Cited by
-
Structure-function analysis of Plasmodium RNA triphosphatase and description of a triphosphate tunnel metalloenzyme superfamily that includes Cet1-like RNA triphosphatases and CYTH proteins.RNA. 2006 Aug;12(8):1468-74. doi: 10.1261/rna.119806. Epub 2006 Jun 29. RNA. 2006. PMID: 16809816 Free PMC article.
-
Polyphosphatase activity of CthTTM, a bacterial triphosphate tunnel metalloenzyme.J Biol Chem. 2008 Nov 7;283(45):31047-57. doi: 10.1074/jbc.M805392200. Epub 2008 Sep 8. J Biol Chem. 2008. PMID: 18782773 Free PMC article.
-
Mutational analysis of a helicase motif-based RNA 5'-triphosphatase/NTPase from bamboo mosaic virus.Virology. 2007 Oct 10;367(1):41-50. doi: 10.1016/j.virol.2007.05.013. Epub 2007 Jun 22. Virology. 2007. PMID: 17585982 Free PMC article.
-
Magnesium-binding studies reveal fundamental differences between closely related RNA triphosphatases.Nucleic Acids Res. 2008 Feb;36(2):451-61. doi: 10.1093/nar/gkm1067. Epub 2007 Nov 26. Nucleic Acids Res. 2008. PMID: 18039706 Free PMC article.
-
Structural Determinants for Substrate Binding and Catalysis in Triphosphate Tunnel Metalloenzymes.J Biol Chem. 2015 Sep 18;290(38):23348-60. doi: 10.1074/jbc.M115.674473. Epub 2015 Jul 28. J Biol Chem. 2015. PMID: 26221030 Free PMC article.
References
-
- Shuman S. (2001) The mRNA capping apparatus as drug target and guide to eukaryotic phylogeny. Cold Spring Harb. Symp. Quant. Biol., 66, 301–312. - PubMed
-
- Shuman S. (2002) What mRNA capping tells us about eukaryotic evolution. Nature Rev. Mol. Cell Biol., 3, 619–625. - PubMed
-
- Herniou E.A., Olszewski,J.A., Cory,J.S. and O’Reilly,D.R. (2003) The genome sequence and evolution of baculoviruses. Annu. Rev. Entomol., 48, 211–234. - PubMed

