New restriction enzymes discovered from Escherichia coli clinical strains using a plasmid transformation method
- PMID: 12595571
- PMCID: PMC149844
- DOI: 10.1093/nar/gng022
New restriction enzymes discovered from Escherichia coli clinical strains using a plasmid transformation method
Abstract
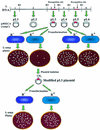
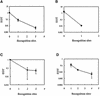
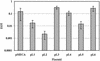
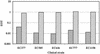
The presence of restriction enzymes in bacterial cells has been predicted by either classical phage restriction-modification (R-M) tests, direct in vitro enzyme assays or more recently from bacterial genome sequence analysis. We have applied phage R-M test principles to the transformation of plasmid DNA and established a plasmid R-M test. To validate this test, six plasmids that contain BamHI fragments of phage lambda DNA were constructed and transformed into Escherichia coli strains containing known R-M systems including: type I (EcoBI, EcoAI, Eco124I), type II (HindIII) and type III (EcoP1I). Plasmid DNA with a single recognition site showed a reduction of relative efficiency of transformation (EOT = 10(-1)-10(-2)). When multiple recognition sites were present, greater reductions in EOT values were observed. Once established in the cell, the plasmids were subjected to modification (EOT = 1.0). We applied this test to screen E.coli clinical strains and detected the presence of restriction enzymes in 93% (14/15) of cells. Using additional subclones and the computer program, RM Search, we identified four new restriction enzymes, Eco377I, Eco585I, Eco646I and Eco777I, along with their recognition sequences, GGA(8N)ATGC, GCC(6N)TGCG, CCA(7N)CTTC, and GGA(6N)TATC, respectively. Eco1158I, an isoschizomer of EcoBI, was also found in this study.
Figures



References
-
- Arber W. and Linn,S. (1969) DNA modification and restriction. Annu. Rev. Biochem., 38, 467–500. - PubMed
-
- Redaschi N. and Bickle,T.A. (1996) DNA restriction and modification systems. In Neidhardt,F.C. (ed.), Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. ASM Press, Washington, DC, pp. 773–781.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

