The role of synaptic GTPase-activating protein in neuronal development and synaptic plasticity
- PMID: 12598599
- PMCID: PMC6742247
- DOI: 10.1523/JNEUROSCI.23-04-01119.2003
The role of synaptic GTPase-activating protein in neuronal development and synaptic plasticity
Abstract
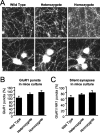
Synaptic GTPase-activating protein (SynGAP) is a neuronal RasGAP (Ras GTPase-activating protein) that is selectively expressed in brain and highly enriched at excitatory synapses, where it negatively regulates Ras activity and its downstream signaling pathways. To investigate the physiological role of SynGAP in the brain, we have generated mutant mice lacking the SynGAP protein. These mice exhibit postnatal lethality, indicating that SynGAP plays a critical role during neuronal development. In addition, cell biological experiments show that neuronal cultures from mutant mice have more synaptic AMPA receptor clusters, suggesting that SynGAP regulates glutamate receptor synaptic targeting. Moreover, electrophysiological studies demonstrated that heterozygous mutant mice have a specific defect in hippocampal long-term potentiation (LTP). These studies show that the regulation of synaptic Ras signaling by SynGAP is important for proper neuronal development and glutamate receptor trafficking and is critical for the induction of LTP.
Figures



References
-
- Bliss TV, Collingridge GL. A synaptic model of memory: long-term potentiation in the hippocampus. Nature. 1993;361:31–39. - PubMed
-
- Bokoch GM, Der CJ. Emerging concepts in the Ras superfamily of GTP-binding proteins. FASEB J. 1993;7:750–759. - PubMed
-
- Brambilla R, Gnesutta N, Minichiello L, White G, Roylance AJ, Herron CE, Ramsey M, Wolfer DP, Cestari V, Rossi-Arnaud C, Grant SG, Chapman PF, Lipp HP, Sturani E, Klein R. A role for the Ras signalling pathway in synaptic transmission and long-term memory. Nature. 1997;390:281–286. - PubMed
-
- Chen HJ, Rojas-Soto M, Oguni A, Kennedy MB. A synaptic Ras-GTPase activating protein (p135 SynGAP) inhibited by CaM kinase II. Neuron. 1998;20:895–904. - PubMed
-
- Dong H, O'Brien RJ, Fung ET, Lanahan AA, Worley PF, Huganir RL. GRIP: a synaptic PDZ domain-containing protein that interacts with AMPA receptors. Nature. 1997;386:279–284. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
