Type I polyketide synthase requiring a discrete acyltransferase for polyketide biosynthesis
- PMID: 12598647
- PMCID: PMC152261
- DOI: 10.1073/pnas.0537286100
Type I polyketide synthase requiring a discrete acyltransferase for polyketide biosynthesis
Abstract
Type I polyketide synthases (PKSs) are multifunctional enzymes that are organized into modules, each of which minimally contains a beta-ketoacyl synthase, an acyltransferase (AT), and an acyl carrier protein. Here we report that the leinamycin (LNM) biosynthetic gene cluster from Streptomyces atroolivaceus S-140 consists of two PKS genes, lnmI and lnmJ, that encode six PKS modules, none of which contain the cognate AT domain. The only AT activity identified within the lnm gene cluster is a discrete AT protein encoded by lnmG. Inactivation of lnmG, lnmI, or lnmJ in vivo abolished LNM biosynthesis. Biochemical characterization of LnmG in vitro showed that it efficiently and specifically loaded malonyl CoA to all six PKS modules. These findings unveiled a previously unknown PKS architecture that is characterized by a discrete, iteratively acting AT protein that loads the extender units in trans to "AT-less" multifunctional type I PKS proteins for polyketide biosynthesis. This PKS structure provides opportunities for PKS engineering as exemplified by overexpressing lnmG to improve LNM production.
Figures
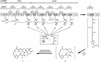
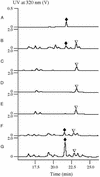


Comment in
-
Polyketide and non-ribosomal peptide synthases: falling together by coming apart.Proc Natl Acad Sci U S A. 2003 Mar 18;100(6):3010-2. doi: 10.1073/pnas.0730689100. Epub 2003 Mar 11. Proc Natl Acad Sci U S A. 2003. PMID: 12631695 Free PMC article. No abstract available.
References
-
- O'Hagan D. Polyketides. Chichester, U.K.: Horwood; 1991.
-
- Hopwood D A. Chem Rev (Washington, DC) 1997;97:2465–2497. - PubMed
-
- Staunton J, Weissman K J. Nat Prod Rep. 2001;18:380–416. - PubMed
-
- Shen B. Top Curr Chem. 2000;209:1–51.
-
- McDaniel R, Ebert-Khosla S, Hopwood D A, Khosla C. Science. 1993;262:1546–1550. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

