SWI/SNF-dependent chromatin remodeling of RNR3 requires TAF(II)s and the general transcription machinery
- PMID: 12600943
- PMCID: PMC195993
- DOI: 10.1101/gad.1039503
SWI/SNF-dependent chromatin remodeling of RNR3 requires TAF(II)s and the general transcription machinery
Abstract
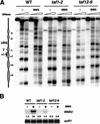
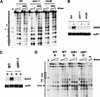
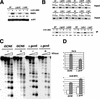
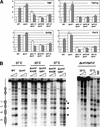
Gene expression requires the recruitment of chromatin remodeling activities and general transcription factors (GTFs) to promoters. Whereas the role of activators in recruiting chromatin remodeling activities has been clearly demonstrated, the contributions of the transcription machinery have not been firmly established. Here we demonstrate that the remodeling of the RNR3 promoter requires a number of GTFs, mediator and RNA polymerase II. We also show that remodeling is dependent upon the SWI/SNF complex, and that TFIID and RNA polymerase II are required for its recruitment to the promoter. In contrast, Gcn5p-dependent histone acetylation occurs independently of TFIID and RNA polymerase II function, and we provide evidence that acetylation increases the extent of nucleosome remodeling, but is not required for SWI/SNF recruitment. Thus, the general transcription machinery can contribute to nucleosome remodeling by mediating the association of SWI/SNF with promoters, thereby revealing a novel pathway for the recruitment of chromatin remodeling activities.
Figures



References
-
- Agalioti T, Lomvardas S, Parekh B, Yie J, Maniatis T, Thanos D. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-β promoter. Cell. 2000;103:667–678. - PubMed
-
- Balasubramanian R, Pray-Grant MG, Selleck W, Grant PA, Tan S. Role of the Ada2 and Ada3 transcriptional coactivators in histone acetylation. J Biol Chem. 2002;277:7989–7995. - PubMed
-
- Belotserkovskaya R, Berger SL. Interplay between chromatin modifying and remodeling complexes in transcriptional regulation. Crit Rev Eukaryot Gene Expr. 1999;9:221–230. - PubMed
-
- Berger SL. Histone modifications in transcriptional regulation. Curr Opin Genet Dev. 2002;12:142–148. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
