The transcriptional activity of human Chromosome 22
- PMID: 12600945
- PMCID: PMC195998
- DOI: 10.1101/gad.1055203
The transcriptional activity of human Chromosome 22
Abstract
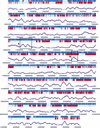
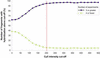
A DNA microarray representing nearly all of the unique sequences of human Chromosome 22 was constructed and used to measure global-transcriptional activity in placental poly(A)(+) RNA. We found that many of the known, related and predicted genes are expressed. More importantly, our study reveals twice as many transcribed bases as have been reported previously. Many of the newly discovered expressed fragments were verified by RNA blot analysis and a novel technique called differential hybridization mapping (DHM). Interestingly, a significant fraction of these novel fragments are expressed antisense to previously annotated introns. The coding potential of these novel expressed regions is supported by their sequence conservation in the mouse genome. This study has greatly increased our understanding of the biological information encoded on a human chromosome. To facilitate the dissemination of these results to the scientific community, we have developed a comprehensive Web resource to present the findings of this study and other features of human Chromosome 22 at http://array.mbb.yale.edu/chr22.
Figures
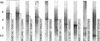


References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Balasubramanian S, Harrison P, Hegyi H, Bertone P, Luscombe N, Echols N, McGarvey P, Zhang Z, Gerstein M. SNPs on human chromosomes 21 and 22—Analysis in terms of protein features and pseudogenes. Pharmacogenomics. 2002;3:393–402. - PubMed
-
- Berman P, Bertone P, DasGupta B, Gerstein M, Kao M-Y, Snyder M. Fast optimal genome tiling with applications to microarray design and homology search. In: Guigo R, Gusfield D, editors. Proceedings of the Second International Workshop on Algorithms in Bioinformatics. 2002. , Lecture Notes in Computer Science, Vol. 2452, pp. 419–433. Springer, Heidelberg.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
