FemABX peptidyl transferases: a link between branched-chain cell wall peptide formation and beta-lactam resistance in gram-positive cocci
- PMID: 12604510
- PMCID: PMC149326
- DOI: 10.1128/AAC.47.3.837-846.2003
FemABX peptidyl transferases: a link between branched-chain cell wall peptide formation and beta-lactam resistance in gram-positive cocci
Figures
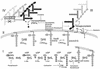
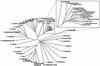
References
-
- Alborn, W. E. J., J. Hoskins, S. Unal, J. E. Flokowitsch, C. A. Hayes, J. E. Dotzlaf, W. K. Yeh, and P. L. Skatrud. 1996. Cloning and characterization of femA and femB from Staphylococcus epidermidis. Gene 180:177-181. - PubMed
-
- al-Obeid, S., D. Billot-Klein, J. van Heijenoort, E. Collatz, and L. Gutmann. 1992. Replacement of the essential penicillin-binding protein 5 by high-molecular mass PBPs may explain vancomycin-beta-lactam synergy in low-level vancomycin-resistant Enterococcus faecium D366. FEMS Microbiol. Lett. 70:79-84. - PubMed
-
- Baba, T., F. Takeuchi, M. Kuroda, H. Yuzawa, K. Aoki, A. Oguchi, Y. Nagai, N. Iwama, K. Asano, T. Naimi, H. Kuroda, L. Cui, K. Yamamoto, and K. Hiramatsu. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359:1819-1827. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

