Quinolone-DNA interaction: sequence-dependent binding to single-stranded DNA reflects the interaction within the gyrase-DNA complex
- PMID: 12604512
- PMCID: PMC149327
- DOI: 10.1128/AAC.47.3.854-862.2003
Quinolone-DNA interaction: sequence-dependent binding to single-stranded DNA reflects the interaction within the gyrase-DNA complex
Abstract
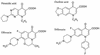
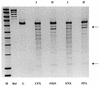
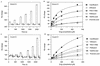
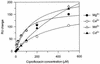
We have investigated the interaction of quinolones with DNA by a number of methods to establish whether a particular binding mode correlates with quinolone potency. The specificities of the quinolone-mediated DNA cleavage reaction of DNA gyrase were compared for a number of quinolones. Two patterns that depended on the potency of the quinolone were identified. Binding to plasmid DNA was examined by measuring the unwinding of pBR322 by quinolones; no correlation with quinolone potency was observed. Quinolone binding to short DNA oligonucleotides was measured by surface plasmon resonance. The quinolones bound to both single- and double-stranded oligonucleotides in an Mg(2+)-dependent manner. Quinolones bound to single-stranded DNA with a higher affinity, and the binding exhibited sequence dependence; binding to double-stranded DNA was sequence independent. The variations in binding in the presence of metal ions showed that Mg(2+) promoted tighter, more specific binding to single-stranded DNA than softer metal ions (Mn(2+) and Cd(2+)). Single-stranded DNA binding by quinolones correlated with the in vitro quinolone potency, indicating that this mode of interaction may reflect the interaction of the quinolone with DNA in the context of the gyrase-DNA complex.
Figures

References
-
- Bailly, C., C. Tardy, L. Wang, B. Armitage, K. Hopkins, A. Kumar, G. B. Schuster, D. W. Boykin, and W. D. Wilson. 2001. Recognition of ATGA sequences by the unfused aromatic dication DB293 forming stacked dimers in the DNA minor groove. Biochemistry 40:9770-9779. - PubMed
-
- Bates, A. D., and A. Maxwell. 1993. DNA topology. IRL Press, Oxford, United Kingdom.
-
- Bhanot, S. K., M. Singh, and N. R. Chatterjee. 2001. The chemical and biological aspects of fluoroquinolones: reality and dreams. Curr. Pharm. Des. 7:311-335. - PubMed
-
- Carrasco, C., H. Vezin, W. D. Wilson, J. Ren, J. B. Chaires, and C. Bailly. 2001. DNA binding properties of the indolocarbazole antitumor drug NB-506. Anticancer Drug Des. 16:99-107. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

