von Hippel-Lindau protein binds hyperphosphorylated large subunit of RNA polymerase II through a proline hydroxylation motif and targets it for ubiquitination
- PMID: 12604794
- PMCID: PMC151405
- DOI: 10.1073/pnas.0436037100
von Hippel-Lindau protein binds hyperphosphorylated large subunit of RNA polymerase II through a proline hydroxylation motif and targets it for ubiquitination
Abstract
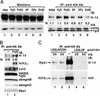
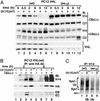
The transition from transcription initiation to elongation involves phosphorylation of the large subunit (Rpb1) of RNA polymerase II on the repetitive carboxyl-terminal domain. The elongating hyperphosphorylated Rpb1 is subject to ubiquitination, particularly in response to UV radiation and DNA-damaging agents. By using computer modeling, we identified regions of Rpb1 and the adjacent subunit 6 of RNA polymerase II (Rpb6) that share sequence and structural similarity with the domain of hypoxia-inducible transcription factor 1 alpha (HIF-1 alpha) that binds von Hippel-Lindau tumor suppressor protein (pVHL). pVHL confers substrate specificity to the E3 ligase complex, which ubiquitinates HIF-alpha and targets it for proteasomal degradation. In agreement with the computational model, we show biochemical evidence that pVHL specifically binds the hyperphosphorylated Rpb1 in a proline-hydroxylation-dependent manner, targeting it for ubiquitination. This interaction is regulated by UV radiation.
Figures




References
-
- Kibel A, Iliopoulos O, DeCaprio J A, Kaelin W G., Jr Science. 1995;269:1444–1446. - PubMed
-
- Kamura T, Koepp D M, Conrad M N, Skowyra D, Moreland R J, Iliopoulos O, Lane W S, Kaelin W G, Jr, Elledge S J, Conaway R C, et al. Science. 1999;284:657–661. - PubMed
-
- Cockman M E, Masson N, Mole D R, Jaakola P, Chang G-W, Clifford S C, Maher E R, Pugh C W, Ratcliffe P J, Maxwell P H. J Biol Chem. 2000;275:25733–25741. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

