A role for N-glycanase in the cytosolic turnover of glycoproteins
- PMID: 12606569
- PMCID: PMC150340
- DOI: 10.1093/emboj/cdg107
A role for N-glycanase in the cytosolic turnover of glycoproteins
Abstract
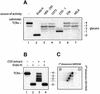
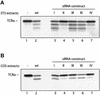
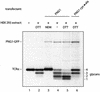
Successful maturation determines the intracellular fate of secretory and membrane proteins in the endoplasmic reticulum (ER). Failure of proteins to fold or assemble properly can lead to their retention in the ER and redirects them to the cytosol for degradation by the proteasome. Proteasome inhibitors can yield deglycosylated cytoplasmic intermediates that are the result of an N-glycanase activity, believed to act prior to destruction of these substrates by the proteasome. A gene encoding a yeast peptide:N-glycanase, PNG1, has been cloned, but this N-glycanase and its mammalian homolog were reported to be incapable of deglycosylating full-length glycoproteins. We show that both the yeast PNG1 enzyme and its mammalian homolog display N-glycanase activity towards intact glycoproteins. As substrates, cytosolic PNGase activity prefers proteins containing high-mannose over those bearing complex type oligosaccharides. Importantly, PNG1 discriminates between non-native and folded glycoproteins, consistent with a role for N-glycanase in cytoplasmic turnover of glycoproteins.
Figures






References
-
- Bacik I. et al. (1997) Introduction of a glycosylation site into a secreted protein provides evidence for an alternative antigen processing pathway: transport of precursors of major histocompatibility complex class I-restricted peptides from the endoplasmic reticulum to the cytosol. J. Exp. Med., 186, 479–487. - PMC - PubMed
-
- Barber L.D. et al. (1996) Unusual uniformity of the N-linked oligosaccharides of HLA-A, -B and -C glycoproteins. J. Immunol., 156, 3275–3284. - PubMed
-
- Barnstable C.J., Bodmer,W.F., Brown,G., Galfre,G., Milstein,C., Williams,A.F. and Ziegler,A. (1978) Production of monoclonal antibodies to group A erythrocytes, HLA and other human cell surface antigens—new tools for genetic analysis. Cell, 14, 9–20. - PubMed
-
- Baumeister W., Walz,J., Zuhl,F. and Seemuller,E. (1998) The proteasome: paradigm of a self-compartmentalizing protease. Cell, 92, 367–380. - PubMed
-
- Brummelkamp T.R., Bernards,R. and Agami,R. (2002) A system for stable expression of short interfering RNAs in mammalian cells. Science, 296, 550–553. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

