Differentiation, cell fusion, and nuclear fusion during ex vivo repair of epithelium by human adult stem cells from bone marrow stroma
- PMID: 12606728
- PMCID: PMC151352
- DOI: 10.1073/pnas.0437997100
Differentiation, cell fusion, and nuclear fusion during ex vivo repair of epithelium by human adult stem cells from bone marrow stroma
Abstract
To investigate stem cell differentiation in response to tissue injury, human mesenchymal stem cells (hMSCs) were cocultured with heat-shocked small airway epithelial cells. A subset of the hMSCs rapidly differentiated into epithelium-like cells, and they restored the epithelial monolayer. Immunocytochemistry and microarray analyses demonstrated that the cells expressed many genes characteristic of normal small airway epithelial cells. Some hMSCs differentiated directly after incorporation into the epithelial monolayer but other hMSCs fused with epithelial cells. Surprisingly, cell fusion was a frequent rather than rare event, in that up to 1% of the hMSCs added to the coculture system were recovered as binucleated cells expressing an epithelial surface epitope. Some of the fused cells also underwent nuclear fusion.
Figures
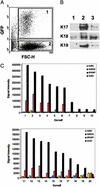

References
-
- Davidson J M. In: Inflammation: Basic Principles and Clinical Correlates. 2nd Ed. Gallin J I, Goldstein I M, Synderman R, editors. New York: Raven; 1992. p. 809.
-
- Gage F H. Science. 2000;287:1433–1438. - PubMed
-
- Alison M R, Poulsom R, Forbes S J. Liver. 2001;21:367–373. - PubMed
-
- Doherty M J, Ashton B A, Walsh S, Beresford J N, Grant M E, Canfield A E. J Bone Miner Res. 1998;13:828–838. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

