Retrofitting BACs with G418 resistance, luciferase, and oriP and EBNA-1 - new vectors for in vitro and in vivo delivery
- PMID: 12609052
- PMCID: PMC150596
- DOI: 10.1186/1472-6750-3-2
Retrofitting BACs with G418 resistance, luciferase, and oriP and EBNA-1 - new vectors for in vitro and in vivo delivery
Abstract
Background: Bacterial artificial chromosomes (BACs) have been used extensively for sequencing the human and mouse genomes and are thus readily available for most genes. The large size of BACs means that they can generally carry intact genes with all the long range controlling elements that drive full levels of tissue-specific expression. For gene expression studies and gene therapy applications it is useful to be able to retrofit the BACs with selectable genes such as G418 resistance, reporter genes such as luciferase, and oriP/EBNA-1 from Epstein Barr virus which allows long term episomal maintenance in mammalian cells.
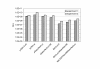
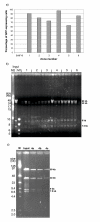
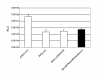
Results: We describe a series of retrofitting plasmids and a protocol for in vivo loxP/Cre recombination. The vector pRetroNeo carries a G418 resistance cassette, pRetroNeoLuc carries G418 resistance and a luciferase expression cassette, pRetroNeoLucOE carries G418 resistance, luciferase and an oriP/EBNA-1 cassette and pRetroNeoOE carries G418 resistance and oriP/EBNA-1. These vectors can be efficiently retrofitted onto BACs without rearrangement of the BAC clone. The luciferase cassette is expressed efficiently from the retrofitting plasmids and from retrofitted BACs after transient transfection of B16F10 cells in tissue culture and after electroporation into muscles of BALB/c mice in vivo. We also show that a BAC carrying GFP, oriP and EBNA-1 can be transfected into B16F10 cells with Lipofectamine 2000 and can be rescued intact after 5 weeks.
Conclusion: The pRetro vectors allow efficient retrofitting of BACs with G418 resistance, luciferase and/or oriP/EBNA-1 using in vivo expression of Cre. The luciferase reporter gene is expressed after transient transfection of retrofitted BACs into cells in tissue culture and after electroporation into mouse muscle in vivo. OriP/EBNA-1 allows stable maintenance of a 150-kb BAC without rearrangement for at least 5 weeks.
Figures


Similar articles
-
In vitro and in vivo delivery of intact BAC DNA -- comparison of different methods.J Gene Med. 2004 Feb;6(2):195-209. doi: 10.1002/jgm.481. J Gene Med. 2004. PMID: 14978773
-
Enhancement of transgene expression by cotransfection of oriP plasmid with EBNA-1 expression vector.Hum Gene Ther. 2000 Feb 10;11(3):471-9. doi: 10.1089/10430340050015932. Hum Gene Ther. 2000. PMID: 10697121
-
Factor VIII mRNA expression from a BAC carrying the intact locus made by homologous recombination.Genomics. 2007 Nov;90(5):610-9. doi: 10.1016/j.ygeno.2007.07.005. Epub 2007 Sep 5. Genomics. 2007. PMID: 17822869
-
Viral bacterial artificial chromosomes: generation, mutagenesis, and removal of mini-F sequences.J Biomed Biotechnol. 2012;2012:472537. doi: 10.1155/2012/472537. Epub 2012 Feb 23. J Biomed Biotechnol. 2012. PMID: 22496607 Free PMC article. Review.
-
Applications of muscle electroporation gene therapy.Curr Gene Ther. 2002 Feb;2(1):101-5. doi: 10.2174/1566523023348183. Curr Gene Ther. 2002. PMID: 12108971 Review.
Cited by
-
Cross-species RNAi rescue platform in Drosophila melanogaster.Genetics. 2009 Nov;183(3):1165-73. doi: 10.1534/genetics.109.106567. Epub 2009 Aug 31. Genetics. 2009. PMID: 19720858 Free PMC article.
-
Transcriptional Suppression of Diabetic Nephropathy with Novel Gene Silencer Pyrrole-Imidazole Polyamides Preventing USF1 Binding to the TGF-β1 Promoter.Int J Mol Sci. 2021 Apr 29;22(9):4741. doi: 10.3390/ijms22094741. Int J Mol Sci. 2021. PMID: 33947045 Free PMC article.
-
Bacterial delivery of large intact genomic-DNA-containing BACs into mammalian cells.Bioeng Bugs. 2012 Mar-Apr;3(2):86-92. doi: 10.4161/bbug.18621. Epub 2012 Mar 1. Bioeng Bugs. 2012. PMID: 22095052 Free PMC article.
-
Optimizing targeted gene delivery: chemical modification of viral vectors and synthesis of artificial virus vector systems.AAPS J. 2006;8(4):E731-42. doi: 10.1208/aapsj080483. AAPS J. 2006. PMID: 17285739 Free PMC article. Review.
-
Recombining overlapping BACs into a single larger BAC.BMC Biotechnol. 2004 Jan 6;4:1. doi: 10.1186/1472-6750-4-1. BMC Biotechnol. 2004. PMID: 14709179 Free PMC article.
References
-
- Yang XW, Model P, Heintz N. Homologous recombination based modification in Esherichia coli and germline transmission in transgenic mice of a bacterial artificial chromosome. Nature Biotechnology. 1997;15:859–865. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

