MD simulation of protein-ligand interaction: formation and dissociation of an insulin-phenol complex
- PMID: 12609856
- PMCID: PMC1302723
- DOI: 10.1016/S0006-3495(03)74962-5
MD simulation of protein-ligand interaction: formation and dissociation of an insulin-phenol complex
Abstract
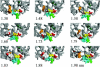
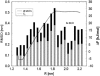
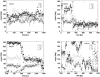
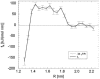
Complexes of proteins with small ligands are of utmost importance in biochemistry, and therefore equilibria, formation, and decay have been investigated extensively by means of biochemical and biophysical methods. Theoretical studies of the molecular dynamics of such systems in solution are restricted to 10 ns, i.e., to fast processes. Only recently new theoretical methods have been developed not to observe the process in real time, but to explore its pathway(s) through the energy landscape. From the profiles of free energy, equilibrium and kinetic quantities can be determined using transition-state theory. This study is dedicated to the pharmacologically relevant insulin-phenol complex. The distance of the center of mass chosen as a reaction coordinate allows a reasonable description over most of the pathway. The analysis is facilitated by analytical expressions we recently derived for distance-type reaction coordinates. Only the sudden onset of rotations at the very release of the ligand cannot be parameterized by a distance. They obviously require a particular treatment. Like a preliminary study on a peptide, the present case emphasizes the contribution of internal friction inside a protein, which can be computed from simulation data. The calculated equilibrium constant and the friction-corrected rates agree well with experimental data.
Figures








Similar articles
-
Ligand escape pathways and (un)binding free energy calculations for the hexameric insulin-phenol complex.Biophys J. 2008 Nov 1;95(9):4193-204. doi: 10.1529/biophysj.108.139675. Epub 2008 Aug 1. Biophys J. 2008. PMID: 18676643 Free PMC article.
-
Phenol-benzene complexation dynamics: quantum chemistry calculation, molecular dynamics simulations, and two dimensional IR spectroscopy.J Chem Phys. 2006 Dec 28;125(24):244508. doi: 10.1063/1.2403132. J Chem Phys. 2006. PMID: 17199356
-
Coordinate and time-dependent diffusion dynamics in protein folding.Methods. 2010 Sep;52(1):91-8. doi: 10.1016/j.ymeth.2010.04.016. Epub 2010 May 11. Methods. 2010. PMID: 20438841 Review.
-
Nonequilibrium, multiple-timescale simulations of ligand-receptor interactions in structured protein systems.Proteins. 2003 Aug 15;52(3):339-48. doi: 10.1002/prot.10411. Proteins. 2003. PMID: 12866048
-
Towards understanding the mechanisms of molecular recognition by computer simulations of ligand-protein interactions.J Mol Recognit. 1999 Nov-Dec;12(6):371-89. doi: 10.1002/(SICI)1099-1352(199911/12)12:6<371::AID-JMR479>3.0.CO;2-O. J Mol Recognit. 1999. PMID: 10611647 Review.
Cited by
-
Structure-affinity properties of a high-affinity ligand of FKBP12 studied by molecular simulations of a binding intermediate.PLoS One. 2014 Dec 12;9(12):e114610. doi: 10.1371/journal.pone.0114610. eCollection 2014. PLoS One. 2014. PMID: 25502559 Free PMC article.
-
The structures of the E22Δ mutant-type amyloid-β alloforms and the impact of E22Δ mutation on the structures of the wild-type amyloid-β alloforms.ACS Chem Neurosci. 2013 Feb 20;4(2):310-20. doi: 10.1021/cn300149j. Epub 2012 Dec 18. ACS Chem Neurosci. 2013. PMID: 23421682 Free PMC article.
-
Small-Molecule Inhibitors of TIPE3 Protein Identified through Deep Learning Suppress Cancer Cell Growth In Vitro.Cells. 2024 Apr 30;13(9):771. doi: 10.3390/cells13090771. Cells. 2024. PMID: 38727307 Free PMC article.
-
Interactions Between Peptide and Preservatives: Effects on Peptide Self-Interactions and Antimicrobial Efficiency In Aqueous Multi-Dose Formulations.Pharm Res. 2015 Oct;32(10):3201-12. doi: 10.1007/s11095-015-1697-z. Epub 2015 Apr 18. Pharm Res. 2015. PMID: 25893328
-
Mechanisms for Benzene Dissociation through the Excited State of T4 Lysozyme L99A Mutant.Biophys J. 2019 Jan 22;116(2):205-214. doi: 10.1016/j.bpj.2018.09.035. Epub 2018 Dec 8. Biophys J. 2019. PMID: 30606449 Free PMC article.
References
-
- Baker, E. N., T. L. Blundell, J. F. Cutfield, S. M. Cutfield, E. J. Dodson, G. G. Dodson, D. M. Crowfoot Hodgkin, R. E. Hubbard, N. W. Isaacs, C. D. Reynolds, K. Sakabe, N. Sakabe, and N. M. Vijayan. 1988. The structure of 2Zn pig insulin crystals at 1.5 Å resolution. Philos. Trans. R. Soc. Lond. B. 319:369–456. - PubMed
-
- Bentley, G., E. Dodson, G. Dodson, D. Hodgkin, and D. Mercola. 1976. Structure of insulin in 4-zinc insulin. Nature. 261:166–168. - PubMed
-
- Berchtold, H., and R. Hilgenfeld. 1999. Binding of phenol to R6 insulin hexamers. Biopolymers. 51:165–172. - PubMed
-
- Birnbaum, D. T., M. A. Kilcomons, M. R. DeFelippis, and J. M. Beals. 1997. Assembly and dissociation of human insulin and LysB28ProB29-insulin hexamers: a comparison study. Pharm. Res. 14:25–36. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

