Force generation by actin polymerization II: the elastic ratchet and tethered filaments
- PMID: 12609863
- PMCID: PMC1302730
- DOI: 10.1016/S0006-3495(03)74969-8
Force generation by actin polymerization II: the elastic ratchet and tethered filaments
Abstract
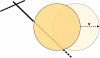
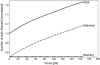
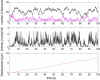
The motion of many intracellular pathogens is driven by the polymerization of actin filaments. The propulsive force developed by the polymerization process is thought to arise from the thermal motions of the polymerizing filament tips. Recent experiments suggest that the nucleation of actin filaments involves a phase when the filaments are attached to the pathogen surface by a protein complex. Here we extend the "elastic ratchet model" of Mogilner and Oster to incorporate these new findings. We apply this "tethered ratchet" model to derive the force-velocity relation for Listeria and discuss relations of our theoretical predictions to experimental measurements. We also discuss "symmetry breaking" dynamics observed in ActA-coated bead experiments, and the implications of the model for lamellipodial protrusion in migrating cells.
Figures





Similar articles
-
Soft Listeria: actin-based propulsion of liquid drops.Phys Rev E Stat Nonlin Soft Matter Phys. 2004 Jun;69(6 Pt 1):061906. doi: 10.1103/PhysRevE.69.061906. Epub 2004 Jun 2. Phys Rev E Stat Nonlin Soft Matter Phys. 2004. PMID: 15244616
-
Mechanics model for actin-based motility.Phys Rev E Stat Nonlin Soft Matter Phys. 2009 Feb;79(2 Pt 1):021916. doi: 10.1103/PhysRevE.79.021916. Epub 2009 Feb 26. Phys Rev E Stat Nonlin Soft Matter Phys. 2009. PMID: 19391787
-
Cooperative symmetry-breaking by actin polymerization in a model for cell motility.Nat Cell Biol. 1999 Dec;1(8):493-9. doi: 10.1038/70281. Nat Cell Biol. 1999. PMID: 10587645
-
Actin cytoskeleton: the Arp2/3 complex gets to the point.Curr Biol. 1998 Sep 10;8(18):R654-7. doi: 10.1016/s0960-9822(07)00415-0. Curr Biol. 1998. PMID: 9740796 Review.
-
On the edge: modeling protrusion.Curr Opin Cell Biol. 2006 Feb;18(1):32-9. doi: 10.1016/j.ceb.2005.11.001. Epub 2005 Nov 28. Curr Opin Cell Biol. 2006. PMID: 16318917 Review.
Cited by
-
The role of membrane stiffness and actin turnover on the force exerted by DRG lamellipodia.Biophys J. 2012 Jun 6;102(11):2451-60. doi: 10.1016/j.bpj.2012.04.036. Epub 2012 Jun 5. Biophys J. 2012. PMID: 22713560 Free PMC article.
-
Analysis of barotactic and chemotactic guidance cues on directional decision-making of Dictyostelium discoideum cells in confined environments.Proc Natl Acad Sci U S A. 2020 Oct 13;117(41):25553-25559. doi: 10.1073/pnas.2000686117. Epub 2020 Sep 30. Proc Natl Acad Sci U S A. 2020. PMID: 32999070 Free PMC article.
-
Crawling cell locomotion revisited.Proc Natl Acad Sci U S A. 2011 Dec 20;108(51):20275-6. doi: 10.1073/pnas.1116814108. Epub 2011 Dec 9. Proc Natl Acad Sci U S A. 2011. PMID: 22159034 Free PMC article. No abstract available.
-
Mathematical modeling of eukaryotic cell migration: insights beyond experiments.Annu Rev Cell Dev Biol. 2013;29:501-28. doi: 10.1146/annurev-cellbio-101512-122308. Epub 2013 Jul 24. Annu Rev Cell Dev Biol. 2013. PMID: 23909278 Free PMC article. Review.
-
Adaptive nonequilibrium design of actin-based metamaterials: Fundamental and practical limits of control.Proc Natl Acad Sci U S A. 2024 Feb 20;121(8):e2310238121. doi: 10.1073/pnas.2310238121. Epub 2024 Feb 15. Proc Natl Acad Sci U S A. 2024. PMID: 38359294 Free PMC article.
References
-
- Bear, J. E., M. Krause, and F. B. Gertler. 2001. Regulating cellular actin assembly. Curr. Opin. Cell Biol. 13:158–166. - PubMed
-
- Beckerle, M. C. 1998. Spatial control of actin filament assembly: lessons from Listeria. Cell. 95:741–748. - PubMed
-
- Bernheim-Groswasser, A., S. Wiesne, R. Golsteyn, M. F. Carlier, and C. Sykes. 2002. The dynamics of actin-based motility depend on surface parameters. Nature. 417:308–311. - PubMed
-
- Borisy, G. G., and T. M. Svitkina. 2000. Actin machinery: pushing the envelope. Curr. Opin. Cell Biol. 12:104–112. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

