Short peptide amyloid organization: stabilities and conformations of the islet amyloid peptide NFGAIL
- PMID: 12609890
- PMCID: PMC1302757
- DOI: 10.1016/S0006-3495(03)74996-0
Short peptide amyloid organization: stabilities and conformations of the islet amyloid peptide NFGAIL
Abstract
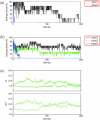
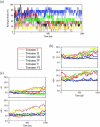
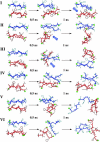
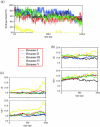
Experimentally, short peptides have been shown to form amyloids similar to those of their parent proteins. Consequently, they present useful systems for studies of amyloid conformation. Here we simulate extensively the NFGAIL peptide, derived from the human islet amyloid polypeptide (residues 22-27). We simulate different possible strand/sheet organizations, from dimers to nonamers. Our simulations indicate that the most stable conformation is an antiparallel strand orientation within the sheets and parallel between sheets. Consistent with the alanine mutagenesis, we find that the driving force is the hydrophobic effect. Whereas the NFGAIL forms stable oligomers, the NAGAIL oligomer is unstable, and disintegrates very quickly after the beginning of the simulation. The simulations further identify a minimal seed size. Combined with our previous simulations of the prion-derived AGAAAAGA peptide, AAAAAAAA, and the Alzheimer Abeta fragments 16-22, 24-36, 16-35, and 10-35, and the solid-state NMR data for Abeta fragments 16-22, 10-35, and 1-40, some insight into the length and the sequence matching effects may be obtained.
Figures



Similar articles
-
The role of Phe in the formation of well-ordered oligomers of amyloidogenic hexapeptide (NFGAIL) observed in molecular dynamics simulations with explicit solvent.Biophys J. 2005 Apr;88(4):2897-906. doi: 10.1529/biophysj.104.055574. Epub 2005 Jan 14. Biophys J. 2005. PMID: 15653723 Free PMC article.
-
Molecular dynamics simulations of alanine rich beta-sheet oligomers: Insight into amyloid formation.Protein Sci. 2002 Oct;11(10):2335-50. doi: 10.1110/ps.4270102. Protein Sci. 2002. PMID: 12237456 Free PMC article.
-
Stabilities and conformations of Alzheimer's beta -amyloid peptide oligomers (Abeta 16-22, Abeta 16-35, and Abeta 10-35): Sequence effects.Proc Natl Acad Sci U S A. 2002 Oct 29;99(22):14126-31. doi: 10.1073/pnas.212206899. Epub 2002 Oct 21. Proc Natl Acad Sci U S A. 2002. PMID: 12391326 Free PMC article.
-
Understanding amyloid fibril nucleation and aβ oligomer/drug interactions from computer simulations.Acc Chem Res. 2014 Feb 18;47(2):603-11. doi: 10.1021/ar4002075. Epub 2013 Dec 24. Acc Chem Res. 2014. PMID: 24368046 Review.
-
Molecular structures of amyloid and prion fibrils: consensus versus controversy.Acc Chem Res. 2013 Jul 16;46(7):1487-96. doi: 10.1021/ar300282r. Epub 2013 Jan 7. Acc Chem Res. 2013. PMID: 23294335 Free PMC article. Review.
Cited by
-
Protein aggregation: in silico algorithms and applications.Biophys Rev. 2021 Jan 17;13(1):71-89. doi: 10.1007/s12551-021-00778-w. eCollection 2021 Feb. Biophys Rev. 2021. PMID: 33747245 Free PMC article. Review.
-
Sampling the self-assembly pathways of KFFE hexamers.Biophys J. 2004 Dec;87(6):3648-56. doi: 10.1529/biophysj.104.047688. Epub 2004 Sep 17. Biophys J. 2004. PMID: 15377527 Free PMC article.
-
The stability and dynamics of the human calcitonin amyloid peptide DFNKF.Biophys J. 2004 Jul;87(1):146-58. doi: 10.1529/biophysj.104.040352. Biophys J. 2004. PMID: 15240453 Free PMC article.
-
Mechanisms for the inhibition of amyloid aggregation by small ligands.Biosci Rep. 2016 Sep 29;36(5):e00385. doi: 10.1042/BSR20160101. Print 2016 Oct. Biosci Rep. 2016. PMID: 27512096 Free PMC article.
-
Zn2+-triggered self-assembly of Gonadorelin [6-D-Phe] to produce nanostructures and fibrils.Sci Rep. 2018 Jul 26;8(1):11280. doi: 10.1038/s41598-018-29529-w. Sci Rep. 2018. PMID: 30050082 Free PMC article.
References
-
- Azriel, R., and E. Gazit. 2001. Analysis of the minimal amyloid-forming fragment of the islet amyloid polypeptide. J. Biol. Chem. 276:34156–34161. - PubMed
-
- Balbach, J. J., Y. Ishii, N. O. Oleg, R. D. Leapman, N. W. Rizzo, F. Dyda, J. Reed, and R. Tykco. 2000. Amyloid fibril formation by Aβ16–22: a seven-residue fragment of the Alzheimer's β-amyloid peptide, and structural characterization by solid state NMR. Biochemistry. 39:13748–13759. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

