Triggering of human parainfluenza virus 3 fusion protein (F) by the hemagglutinin-neuraminidase (HN) protein: an HN mutation diminishes the rate of F activation and fusion
- PMID: 12610140
- PMCID: PMC149538
- DOI: 10.1128/jvi.77.6.3647-3654.2003
Triggering of human parainfluenza virus 3 fusion protein (F) by the hemagglutinin-neuraminidase (HN) protein: an HN mutation diminishes the rate of F activation and fusion
Abstract
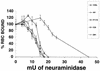
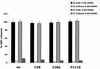
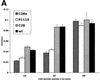
For human parainfluenza virus type 3 and many other paramyxoviruses, membrane fusion mediated by the fusion protein (F) has a stringent requirement for the presence of the homotypic hemagglutinin-neuraminidase protein (HN). With the goal of gaining further insight into the role of HN in the fusion process, we developed a simple method for quantitative comparison of the ability of wild-type and variant HNs to activate F. In this method, HN/F-coexpressing cells with red blood cells (RBC) bound to them at 4 degrees C are transferred to 22 degrees C, and at different times after transfer 4-guanidino-neu5Ac2en (4-GU-DANA) is added; this inhibitor of the HN-receptor interaction then releases all reversibly bound RBC but not those in which F insertion in the target membrane or fusion has occurred. Thus, the amount of irreversibly bound (nonreleased) RBC provides a measure of F activation, and the use of fluorescently labeled RBC permits microscopic assessment of the extent to which F insertion has progressed to fusion. We studied two neuraminidase-deficient HN variants, C28a, which has two mutations, P111S and D216N, and C28, which possesses the D216N mutation only. C28a but not C28 exhibits a slow fusion phenotype, although determination of the HNs' receptor-binding avidity (with our sensitive method, employing RBC with different degrees of receptor depletion) showed that the receptor-binding avidity of C28a or C28 HN was not lower than that of the wild type. The F activation assay, however, revealed fusion-triggering defects in C28a HN. After 10 and also 20 min at 22 degrees C, irreversible RBC binding was significantly less for cells coexpressing wild-type F with C28a HN than for cells coexpressing wild-type F with wild-type HN. In addition, F insertion progressed to fusion more slowly in the case of C28a HN-expressing cells than of wild-type HN-expressing cells. Identical defects were found for P111S HN, whereas for C28 HN, representing the 216 mutation of C28a, F activation and fusion were as rapid as for wild-type HN. The diminished fusion promotion capacity of C28a HN is therefore attributable to P111S, a mutation in the stalk region of the molecule that causes no decrease in receptor-binding avidity. C28a HN is the first parainfluenza virus variant found so far to be specifically defective in HN's F-triggering and fusion promotion functions and may contribute to our understanding of transmission of the activating signal from HN to F.
Figures



References
-
- Bousse, T., T. Takimoto, W. Gorman, T. Takahashi, and A. Portner. 1994. Regions on the hemagglutinin-neuraminidase proteins of human parainfluenza virus type-1 and Sendai virus important for membrane fusion. Virology 204:506-514. - PubMed
-
- Connaris, H., T. Takimoto, R. Russell, S. Crennell, I. Moustafa, A. Portner, and G. Taylor. 2002. Probing the sialic acid binding site of the hemagglutinin-neuraminidase of Newcastle disease virus: identification of key amino acids involved in cell binding, catalysis, and fusion. J. Virol. 76:1816-1824. - PMC - PubMed
-
- Crennell, S., T. Takimoto, A. Portner, and G. Taylor. 2000. Crystal structure of the multifunctional paramyxovirus hemagglutinin-neuraminidase. Nat. Struct. Biol. 7:1068-1074. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

