Role of the virus nucleoprotein in the regulation of lymphocytic choriomeningitis virus transcription and RNA replication
- PMID: 12610166
- PMCID: PMC149515
- DOI: 10.1128/jvi.77.6.3882-3887.2003
Role of the virus nucleoprotein in the regulation of lymphocytic choriomeningitis virus transcription and RNA replication
Abstract
The prototypic arenavirus lymphocytic choriomeningitis virus (LCMV) has a bisegmented negative-strand RNA genome. Each segment carries two viral genes in opposite orientation and separated by an intergenic region (IGR). The RNA-dependent RNA polymerase (RdRp) L of LCMV produces subgenomic mRNA and full-length genomic and antigenomic RNA species in two different processes termed transcription and replication, respectively. It is widely accepted that intracellular nucleoprotein (NP) levels regulate these two processes. Intracellular NP levels increase during the course of the infection, resulting in the unfolding of secondary RNA structures within the IGR. Structure-dependent transcription termination at the IGR is thereby attenuated, promoting replication of genome and antigenome RNA species. To test this hypothesis, we established a helper-virus-free minigenome (MG) system where intracellular synthesis of an S segment analogue from a plasmid is driven by RNA polymerase I. Cotransfection with two additional plasmids expressing the minimal viral trans-acting factors L and NP under control of RNA polymerase II allowed for RNA synthesis mediated by the intracellularly reconstituted LCMV polymerase. Both processes, transcription and replication, were strictly dependent on NP. However, both were equally enhanced by incrementally increasing amounts of NP up to levels in the range of those in LCMV-infected cells. Our data are consistent with a central role for NP in transcription and replication of the LCMV genome, but they do not support the participation of NP levels in balancing the two processes.
Figures

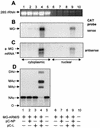

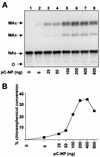
References
-
- Blumberg, B. M., M. Leppert, and D. Kolakofsky. 1981. Interaction of VSV leader RNA and nucleocapsid protein may control VSV genome replication. Cell 23:837-845. - PubMed
-
- Buchmeier, M. J., J. H. Elder, and M. B. Oldstone. 1978. Protein structure of lymphocytic choriomeningitis virus: identification of the virus structural and cell associated polypeptides. Virology 89:133-145. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

