Molecular classification of familial non-BRCA1/BRCA2 breast cancer
- PMID: 12610208
- PMCID: PMC151375
- DOI: 10.1073/pnas.0533805100
Molecular classification of familial non-BRCA1/BRCA2 breast cancer
Abstract
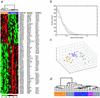
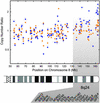
In the decade since their discovery, the two major breast cancer susceptibility genes BRCA1 and BRCA2, have been shown conclusively to be involved in a significant fraction of families segregating breast and ovarian cancer. However, it has become equally clear that a large proportion of families segregating breast cancer alone are not caused by mutations in BRCA1 or BRCA2. Unfortunately, despite intensive effort, the identification of additional breast cancer predisposition genes has so far been unsuccessful, presumably because of genetic heterogeneity, low penetrance, or recessive/polygenic mechanisms. These non-BRCA1/2 breast cancer families (termed BRCAx families) comprise a histopathologically heterogeneous group, further supporting their origin from multiple genetic events. Accordingly, the identification of a method to successfully subdivide BRCAx families into recognizable groups could be of considerable value to further genetic analysis. We have previously shown that global gene expression analysis can identify unique and distinct expression profiles in breast tumors from BRCA1 and BRCA2 mutation carriers. Here we show that gene expression profiling can discover novel classes among BRCAx tumors, and differentiate them from BRCA1 and BRCA2 tumors. Moreover, microarray-based comparative genomic hybridization (CGH) to cDNA arrays revealed specific somatic genetic alterations within the BRCAx subgroups. These findings illustrate that, when gene expression-based classifications are used, BRCAx families can be grouped into homogeneous subsets, thereby potentially increasing the power of conventional genetic analysis.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

