Principal role of TRAP/mediator and SWI/SNF complexes in Kaposi's sarcoma-associated herpesvirus RTA-mediated lytic reactivation
- PMID: 12612078
- PMCID: PMC149486
- DOI: 10.1128/MCB.23.6.2055-2067.2003
Principal role of TRAP/mediator and SWI/SNF complexes in Kaposi's sarcoma-associated herpesvirus RTA-mediated lytic reactivation
Abstract
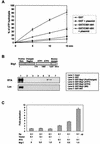
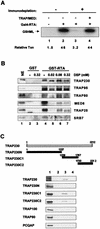
An important step in the herpesvirus life cycle is the switch from latency to lytic reactivation. The RTA transcription activator of Kaposi's sarcoma-associated herpesvirus (KSHV) acts as a molecular switch for lytic reactivation. Here we demonstrate that KSHV RTA recruits CBP, the SWI/SNF chromatin remodeling complex, and the TRAP/Mediator coactivator into viral promoters through interactions with a short acidic sequence in the carboxyl region and that this recruitment is essential for RTA-dependent viral gene expression. The Brg1 subunit of SWI/SNF and the TRAP230 subunit of TRAP/Mediator were shown to interact directly with RTA. Consequently, genetic ablation of these interactions abolished KSHV lytic replication. These results demonstrate that the recruitment of CBP, SWI/SNF, and TRAP/Mediator complexes by RTA is the principal mechanism to direct well-controlled viral gene expression and thereby viral lytic reactivation.
Figures





References
-
- Agalioti, T., S. Lomvardas, B. Parekh, J. Yie, T. Maniatis, and D. Thanos. 2000. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell 103:667-678. - PubMed
-
- Alexander, L., L. Denekamp, A. Knapp, M. R. Auerbach, B. Damania, and R. C. Desrosiers. 2000. The primary sequence of rhesus monkey rhadinovirus isolate 26-95: sequence similarities to Kaposi's sarcoma-associated herpesvirus and rhesus monkey rhadinovirus isolate 17577. J. Virol. 74:3388-3398. - PMC - PubMed
-
- Armstrong, J. A., J. J. Bieker, and B. M. Emerson. 1998. A SWI/SNF-related chromatin remodeling complex, E-RC1, is required for tissue-specific transcriptional regulation by EKLF in vitro. Cell 95:93-104. - PubMed
-
- Boshoff, C., T. F. Schulz, M. M. Kennedy, A. K. Graham, C. Fisher, A. Thomas, J. O. McGee, R. A. Weiss, and J. J. O'Leary. 1995. Kaposi's sarcoma-associated herpesvirus infects endothelial and spindle cells. Nat. Med. 1:1274-1278. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
