Watching proteins fold one molecule at a time
- PMID: 12612345
- PMCID: PMC152269
- DOI: 10.1073/pnas.2628068100
Watching proteins fold one molecule at a time
Erratum in
- Proc Natl Acad Sci U S A. 2003 Jun 10;100(12):7418
Abstract
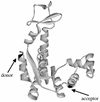
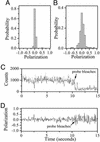
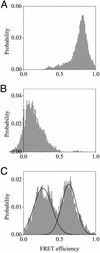
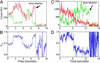
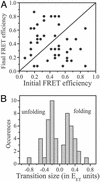
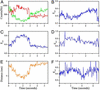
Recent theoretical work suggests that protein folding involves an ensemble of pathways on a rugged energy landscape. We provide direct evidence for heterogeneous folding pathways from single-molecule studies, facilitated by a recently developed immobilization technique. Individual fluorophore-labeled molecules of the protein adenylate kinase were trapped within surface-tethered lipid vesicles, thereby allowing spatial restriction without inducing any spurious interactions with the environment, which often occur when using direct surface-linking techniques. The conformational fluctuations of these protein molecules, prepared at the thermodynamic midtransition point, were studied by using fluorescence resonance energy transfer between two specifically attached labels. Folding and unfolding transitions appeared in experimental time traces as correlated steps in donor and acceptor fluorescence intensity. The size of the steps, in fluorescence resonance energy transfer efficiency units, shows a very broad distribution. This distribution peaks at a relatively low value, indicating a preference for small-step motion on the energy landscape. The time scale of the transitions is also distributed, and although many transitions are too fast to be time-resolved here, the slowest ones may take >1 sec to complete. These extremely slow changes during the folding of single molecules highlight the possible importance of correlated, non-Markovian conformational dynamics.
Figures
Similar articles
-
Fast collapse but slow formation of secondary structure elements in the refolding transition of E. coli adenylate kinase.J Mol Biol. 2005 Sep 23;352(3):683-99. doi: 10.1016/j.jmb.2005.06.074. J Mol Biol. 2005. PMID: 16098987
-
Sequential Closure of Loop Structures Forms the Folding Nucleus during the Refolding Transition of the Escherichia coli Adenylate Kinase Molecule.Biochemistry. 2016 Jan 12;55(1):79-91. doi: 10.1021/acs.biochem.5b00849. Epub 2015 Dec 23. Biochemistry. 2016. PMID: 26666584
-
Deciphering hierarchical features in the energy landscape of adenylate kinase folding/unfolding.J Chem Phys. 2018 Mar 28;148(12):123325. doi: 10.1063/1.5016487. J Chem Phys. 2018. PMID: 29604865
-
The study of protein folding and dynamics by determination of intramolecular distance distributions and their fluctuations using ensemble and single-molecule FRET measurements.Chemphyschem. 2005 May;6(5):858-70. doi: 10.1002/cphc.200400617. Chemphyschem. 2005. PMID: 15884068 Review.
-
Folding funnels and conformational transitions via hinge-bending motions.Cell Biochem Biophys. 1999;31(2):141-64. doi: 10.1007/BF02738169. Cell Biochem Biophys. 1999. PMID: 10593256 Review.
Cited by
-
Real-time submillisecond single-molecule FRET dynamics of freely diffusing molecules with liposome tethering.Nat Commun. 2015 Apr 24;6:6992. doi: 10.1038/ncomms7992. Nat Commun. 2015. PMID: 25908552 Free PMC article.
-
On demand delivery and analysis of single molecules on a programmable nanopore-optofluidic device.Nat Commun. 2019 Aug 16;10(1):3712. doi: 10.1038/s41467-019-11723-7. Nat Commun. 2019. PMID: 31420559 Free PMC article.
-
Orthogonal site-specific protein modification by engineering reversible thiol protection mechanisms.Protein Sci. 2005 Jan;14(1):64-73. doi: 10.1110/ps.04965405. Epub 2004 Dec 2. Protein Sci. 2005. PMID: 15576565 Free PMC article.
-
Microsecond dynamics of an unfolded protein by a line confocal tracking of single molecule fluorescence.Sci Rep. 2013;3:2151. doi: 10.1038/srep02151. Sci Rep. 2013. PMID: 23827883 Free PMC article.
-
A novel immobilization method for single protein spFRET studies.Biophys J. 2005 Aug;89(2):L11-3. doi: 10.1529/biophysj.105.062794. Epub 2005 May 20. Biophys J. 2005. PMID: 15908583 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

