Priming the nucleosome: a role for HMGB proteins?
- PMID: 12612600
- PMCID: PMC1315838
- DOI: 10.1038/sj.embor.embor741
Priming the nucleosome: a role for HMGB proteins?
Abstract
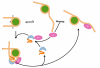
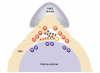
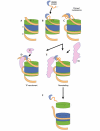
The high-mobility-group B (HMGB) chromosomal proteins are characterized by the HMG box, a DNA-binding domain that both introduces a tight bend into DNA and binds preferentially to a variety of distorted DNA structures. The HMGB proteins seem to act primarily as architectural facilitators in the manipulation of nucleoprotein complexes; for example, in the assembly of complexes involved in recombination and transcription. Recent genetic and biochemical evidence suggests that these proteins can facilitate nucleosome remodelling. One mechanism by which HMGB proteins could prime the nucleosome for migration is to loosen the wrapped DNA and so enhance accessibility to chromatin-remodelling complexes and possibly also to transcription factors. By constraining a tight loop of untwisted DNA at the edge of a nucleosome, an HMGB protein could induce movements in the contacts between certain core histones that would result in an overall change in nucleosome structure.
Figures


References
-
- An W., van Holde K. & Zlatanova J. (1998) The non-histone chromatin protein HMG1 protects linker DNA on the side opposite to that protected by linker histones. J. Biol. Chem., 273, 26289–26291. - PubMed
-
- Anderson J.D., Lowary P.T. & Widom J. (2001) Effects of histone acetylation on the equilibrium accessibility of nucleosomal DNA target sites. J. Mol. Biol., 307, 977–985. - PubMed
-
- Becker P.B. & Hörz W. (2002) ATP-dependent nucleosome remodeling. Annu. Rev. Biochem., 71, 247–273. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

