Infectious hepatitis C virus pseudo-particles containing functional E1-E2 envelope protein complexes
- PMID: 12615904
- PMCID: PMC2193821
- DOI: 10.1084/jem.20021756
Infectious hepatitis C virus pseudo-particles containing functional E1-E2 envelope protein complexes
Abstract
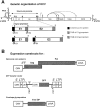
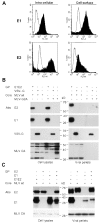
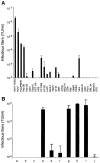
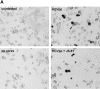
The study of hepatitis C virus (HCV), a major cause of chronic liver disease, has been hampered by the lack of a cell culture system supporting its replication. Here, we have successfully generated infectious pseudo-particles that were assembled by displaying unmodified and functional HCV glycoproteins onto retroviral and lentiviral core particles. The presence of a green fluorescent protein marker gene packaged within these HCV pseudo-particles allowed reliable and fast determination of infectivity mediated by the HCV glycoproteins. Primary hepatocytes as well as hepato-carcinoma cells were found to be the major targets of infection in vitro. High infectivity of the pseudo-particles required both E1 and E2 HCV glycoproteins, and was neutralized by sera from HCV-infected patients and by some anti-E2 monoclonal antibodies. In addition, these pseudo-particles allowed investigation of the role of putative HCV receptors. Although our results tend to confirm their involvement, they provide evidence that neither LDLr nor CD81 is sufficient to mediate HCV cell entry. Altogether, these studies indicate that these pseudo-particles may mimic the early infection steps of parental HCV and will be suitable for the development of much needed new antiviral therapies.
Figures

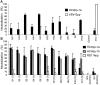
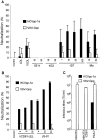
Comment in
-
A model for the study of hepatitis C virus entry.Hepatology. 2003 Sep;38(3):771-4. doi: 10.1002/hep.510380328. Hepatology. 2003. PMID: 12939604
References
-
- Lavanchy, D., R. Purcell, F.B. Hollinger, C. Howard, A. Alberti, M. Kew, G. Dusheiko, M. Alter, E. Ayoola, P. Beutels, et al. 1999. Global surveillance and control of hepatitis C. J. Viral Hepat. 6:35–47. - PubMed
-
- Dieterich, D.T. 2002. Treatment of hepatitis C and anemia in human immunodeficiency virus-infected patients. J. Infect. Dis. 185:S128–137. - PubMed
-
- Lindenbach, B.D., and C.M. Rice. 2001. Flaviviridae: the viruses and their replication. In Fields Virology, 4th ed. D.M. Knipe and P.M. Howley, editors. Lippincott Williams & Wilkins, Philadelphia. 991–1042.
-
- Blight, K.J., A.A. Kolykhalov, and C.M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science. 290:1972–1974. - PubMed
-
- Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 285:110–113. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

