Cell polarity and PIN protein positioning in Arabidopsis require STEROL METHYLTRANSFERASE1 function
- PMID: 12615936
- PMCID: PMC150017
- DOI: 10.1105/tpc.008433
Cell polarity and PIN protein positioning in Arabidopsis require STEROL METHYLTRANSFERASE1 function
Abstract
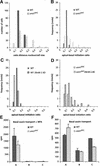
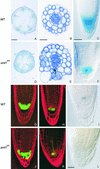
Plants have many polarized cell types, but relatively little is known about the mechanisms that establish polarity. The orc mutant was identified originally by defects in root patterning, and positional cloning revealed that the affected gene encodes STEROL METHYLTRANSFERASE1, which is required for the appropriate synthesis and composition of major membrane sterols. smt1(orc) mutants displayed several conspicuous cell polarity defects. Columella root cap cells revealed perturbed polar positioning of different organelles, and in the smt1(orc) root epidermis, polar initiation of root hairs was more randomized. Polar auxin transport and expression of the auxin reporter DR5-beta-glucuronidase were aberrant in smt1(orc). Patterning defects in smt1(orc) resembled those observed in mutants of the PIN gene family of putative auxin efflux transporters. Consistently, the membrane localization of the PIN1 and PIN3 proteins was disturbed in smt1(orc), whereas polar positioning of the influx carrier AUX1 appeared normal. Our results suggest that balanced sterol composition is a major requirement for cell polarity and auxin efflux in Arabidopsis.
Figures







References
-
- Altmann, T. (1998). Recent advances in brassinosteroid molecular genetics. Curr. Opin. Plant Biol. 1, 378–383. - PubMed
-
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis plants. C. R. Acad. Sci. Paris 316, 1194–1199.
-
- Bligh, E.G., and Dyer, W.J. (1959). A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 91–97. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

