Mutational analysis of the pullulanase-type debranching enzyme of maize indicates multiple functions in starch metabolism
- PMID: 12615940
- PMCID: PMC150021
- DOI: 10.1105/tpc.007575
Mutational analysis of the pullulanase-type debranching enzyme of maize indicates multiple functions in starch metabolism
Abstract
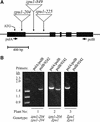
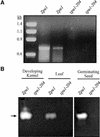
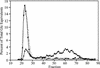
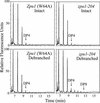
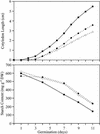
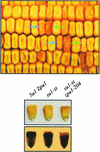
Plants contain two types of alpha(1-->6) glucan hydrolase (starch-debranching enzyme [DBE]). Mutations that affect the pullulanase-type DBE have not been described, although defects in isoamylase-type DBE, known in many plant species, indicate a function in starch biosynthesis. We describe a null mutation of a pullulanase-type DBE gene, a Mutator insertion in maize Zpu1. Plants homozygous for the zpu1-204 mutation are impaired in transient and storage starch degradation. Thus, hydrolytic activity of pullulanase-type DBE contributes to starch catabolism. Developing zpu1-204 endosperm accumulates branched maltooligosaccharides not found in the wild type and is deficient in linear maltooligosaccharides, indicating that the pullulanase-type DBE functions in glucan hydrolysis during kernel starch formation. Furthermore, in a background deficient in isoamylase-type DBE, zpu1-204 conditions a significant accumulation of phytoglycogen in the kernel that is not seen in the wild type. Therefore, pullulanase-type DBE partially compensates for the defect in isoamylase-type DBE, suggesting a function during starch synthesis as well as degradation.
Figures



References
-
- Ball, S., Guan, H.P., James, M., Myers, A., Keeling, P., Mouille, G., Buleon, A., Colonna, P., and Preiss, J. (1996). From glycogen to amylopectin: A model for the biogenesis of the plant starch granule. Cell 86, 349–352. - PubMed
-
- Blauth, S.L., Kim, K.-N., Klucinec, J.D., Shannon, J.C., Thompson, D.B., and Guiltinan, M.J. (2002). Identification of Mutator insertional mutants of starch-branching enzyme 1 (sbe1) in Zea mays L. Plant Mol. Biol. 48, 287–297. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

