A global search reveals epistatic interaction between QTL for early growth in the chicken
- PMID: 12618372
- PMCID: PMC430275
- DOI: 10.1101/gr.528003
A global search reveals epistatic interaction between QTL for early growth in the chicken
Abstract
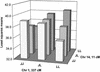
We have identified quantitative trait loci (QTL) explaining a large proportion of the variation in body weights at different ages and growth between chronological ages in an F(2) intercross between red junglefowl and White Leghorn chickens. QTL were mapped using forward selection for loci with significant marginal genetic effects and with a simultaneous search for epistatic QTL pairs. We found 22 significant loci contributing to these traits, nine of these were only found by the simultaneous two-dimensional search, which demonstrates the power of this approach for detecting loci affecting complex traits. We have also estimated the relative contribution of additive, dominance, and epistasis effects to growth and the contribution of epistasis was more pronounced prior to 46 days of age, whereas additive genetic effects explained the major portion of the genetic variance later in life. Several of the detected loci affected either early or late growth but not both. Very few loci affected the entire growth process, which points out that early and late growth, at least to some extent, have different genetic regulation.
Figures


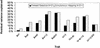
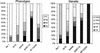
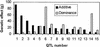
References
-
- Andersson L. 2001. Genetic dissection of phenotypic diversity in farm animals. Nat. Rev. Genet. 2: 130-138. - PubMed
-
- Björnhag G., Knutson, P.-G., and Sperber, I., 1994. Growth–Compendium on the physiology of growth, 3rd ed. (in Swedish). Department of Animal Physiology, Swedish University of Agricultural Sciences, Uppsala, Sweden.
-
- Carlborg Ö. and Andersson, L. 2002. The use of randomization testing for detection of multiple epistatic QTL. Genet. Res. 79: 175-184. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
