Roles of the enantioselective glutathione S-transferases in cleavage of beta-aryl ether
- PMID: 12618439
- PMCID: PMC150126
- DOI: 10.1128/JB.185.6.1768-1775.2003
Roles of the enantioselective glutathione S-transferases in cleavage of beta-aryl ether
Abstract
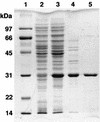
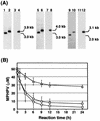
Cleavage of the beta-aryl ether linkage is the most important process in lignin degradation. Here we characterize the three tandemly located glutathione S-transferase (GST) genes, ligF, ligE, and ligG, from low-molecular-weight lignin-degrading Sphingomonas paucimobilis SYK-6, and we describe the actual roles of these genes in the beta-aryl ether cleavage. Based on the identification of the reaction product by electrospray ionization-mass spectrometry, a model compound of beta-aryl ether, alpha-(2-methoxyphenoxy)-beta-hydroxypropiovanillone (MPHPV), was transformed by LigF or LigE to guaiacol and alpha-glutathionyl-beta-hydroxypropiovanillone (GS-HPV). This result suggested that LigF and LigE catalyze the nucleophilic attack of glutathione on the carbon atom at the beta position of MPHPV. High-pressure liquid chromatography-circular dichroism analysis indicated that LigF and LigE each attacked a different enantiomer of the racemic MPHPV preparation. The ligG gene product specifically catalyzed the elimination of glutathione from GS-HPV generated by the action of LigF. This reaction then produces an achiral compound, beta-hydroxypropiovanillone, which is further degraded by this strain. Disruption of the ligF, ligE, and ligG genes in SYK-6 showed that ligF is essential to the degradation of one of the MPHPV enantiomers, and the alternative activities which metabolize the substrates of LigE and LigG are present in this strain.
Figures




References
-
- Adler, E. 1957. Structural elements of lignin. Ind. Eng. Chem. 49:1377-1383.
-
- Adler, E., and E. Eriksoo. 1955. Guaiacylglycerol and its β-guaiacyl ether. Acta Chem. Scand. 9:341-342.
-
- Akiyama, T., K. Magara, Y. Matsumoto, G. Meshitsuka, A. Ishizu, and K. Lundquist. 2000. Proof of the presence of racemic forms of arylglycerol-β-aryl ether structure in lignin: studies on the stereo structure of lignin by ozonation. J. Wood Sci. 46:414-415.
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1990. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
-
- Board, P. G., M. Coggan, G. Chelvanayagam, S. Easteal, L. S. Jermiin, G. K. Schulte, D. E. Danley, L. R. Hoth, M. C. Griffor, A. V. Kamath, M. H. Rosner, B. A. Chrunyk, D. E. Perregaux, C. A. Gabel, K. F. Geoghegan, and J. Pandit. 2000. Identification, characterization, and crystal structure of the omega class glutathione transferases. J. Biol. Chem. 275:24798-24806. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

