Characterization of a novel fucose-regulated promoter (PfcsK) suitable for gene essentiality and antibacterial mode-of-action studies in Streptococcus pneumoniae
- PMID: 12618474
- PMCID: PMC150135
- DOI: 10.1128/JB.185.6.2051-2058.2003
Characterization of a novel fucose-regulated promoter (PfcsK) suitable for gene essentiality and antibacterial mode-of-action studies in Streptococcus pneumoniae
Abstract
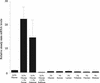
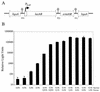
The promoter of the Streptococcus pneumoniae putative fuculose kinase gene (fcsK), the first gene of a novel fucose utilization operon, is induced by fucose and repressed by glucose or sucrose. When the streptococcal polypeptide deformylase (PDF) gene (def1, encoding PDF) was placed under the control of P(fcsK), fucose-dependent growth of the S. pneumoniae (P(fcsK)::def1) strain was observed, confirming the essential nature of PDF in this organism. The mode of antibacterial action of actinonin, a known PDF inhibitor, was also confirmed with this strain. The endogenous fuculose kinase promoter is a tightly regulated, titratable promoter which will be useful for target validation and for confirmation of the mode of action of novel antibacterial drugs in S. pneumoniae.
Figures


References
-
- Chan, P. F., and S. J. Foster. 1998. The role of environmental factors in the regulation of virulence-determinant expression in Staphylococcus aureus 8325-4. Microbiology 144:2469-2479. - PubMed
-
- Chen, D. Z., D. V. Patel, C. J. Hackbarth, W. Wang, G. Dreyer, D. C. Young, P. S. Margolis, C. Wu, Z.-J. Ni, J. Trias, R. J. White, and Z. Yuan. 2000. Actinonin, a naturally occurring antibacterial agent, is a potent deformylase inhibitor. Biochemistry 39:1256-1262. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

