Characterizing the stress/defense transcriptome of Arabidopsis
- PMID: 12620105
- PMCID: PMC153460
- DOI: 10.1186/gb-2003-4-3-r20
Characterizing the stress/defense transcriptome of Arabidopsis
Abstract
Background: To understand the gene networks that underlie plant stress and defense responses, it is necessary to identify and characterize the genes that respond both initially and as the physiological response to the stress or pathogen develops. We used PCR-based suppression subtractive hybridization to identify Arabidopsis genes that are differentially expressed in response to ozone, bacterial and oomycete pathogens and the signaling molecules salicylic acid (SA) and jasmonic acid.
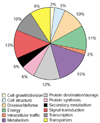
Results: We identified a total of 1,058 differentially expressed genes from eight stress cDNA libraries. Digital northern analysis revealed that 55% of the stress-inducible genes are rarely transcribed in unstressed plants and 17% of them were not previously represented in Arabidopsis expressed sequence tag databases. More than two-thirds of the genes in the stress cDNA collection have not been identified in previous studies as stress/defense response genes. Several stress-responsive cis-elements showed a statistically significant over-representation in the promoters of the genes in the stress cDNA collection. These include W- and G-boxes, the SA-inducible element, the abscisic acid response element and the TGA motif.
Conclusions: The stress cDNA collection comprises a broad repertoire of stress-responsive genes encoding proteins that are involved in both the initial and subsequent stages of the physiological response to abiotic stress and pathogens. This set of stress-, pathogen- and hormone-modulated genes is an important resource for understanding the genetic interactions underlying stress signaling and responses and may contribute to the characterization of the stress transcriptome through the construction of standardized specialized arrays.
Figures




References
-
- Lamb CJ. Plant disease resistance genes in signal perception and transduction. Cell. 1994;76:419–422. - PubMed
-
- Lamb C, Dixon RA. The oxidative burst in plant disease resistance. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:251–275. - PubMed
-
- Baker B, Zambryski P, Staskawicz B, Dinesh-Kumar SP. Signaling in plant-microbe interactions. Science. 1997;276:726–733. - PubMed
-
- Dong X. SA, JA, ethylene, and disease resistance in plants. Curr Opin Plant Biol. 1998;1:316–323. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

