Evolutionary history, structural features and biochemical diversity of the NlpC/P60 superfamily of enzymes
- PMID: 12620121
- PMCID: PMC151301
- DOI: 10.1186/gb-2003-4-2-r11
Evolutionary history, structural features and biochemical diversity of the NlpC/P60 superfamily of enzymes
Abstract
Background: Peptidoglycan is hydrolyzed by a diverse set of enzymes during bacterial growth, development and cell division. The N1pC/P60 proteins define a family of cell-wall peptidases that are widely represented in various bacterial lineages. Currently characterized members are known to hydrolyze D-gamma-glutamyl-meso-diaminopimelate or N-acetylmuramate-L-alanine linkages.
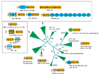
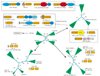
Results: Detailed analysis of the N1pC/P60 peptidases showed that these proteins define a large superfamily encompassing several diverse groups of proteins. In addition to the well characterized P60-like proteins, this superfamily includes the AcmB/LytN and YaeF/YiiX families of bacterial proteins, the amidase domain of bacterial and kinetoplastid glutathionylspermidine synthases (GSPSs), and several proteins from eukaryotes, phages, poxviruses, positive-strand RNA viruses, and certain archaea. The eukaryotic members include lecithin retinol acyltransferase (LRAT), nematode developmental regulator Egl-26, and candidate tumor suppressor H-rev107. These eukaryotic proteins, along with the bacterial YaeF/poxviral G6R family, show a circular permutation of the catalytic domain. We identified three conserved residues, namely a cysteine, a histidine and a polar residue, that are involved in the catalytic activities of this superfamily. Evolutionary analysis of this superfamily shows that it comprises four major families, with diverse domain architectures in each of them.
Conclusions: Several related, but distinct, catalytic activities, such as murein degradation, acyl transfer and amide hydrolysis, have emerged in the N1pC/P60 superfamily. The three conserved catalytic residues of this superfamily are shown to be equivalent to the catalytic triad of the papain-like thiol peptidases. The predicted structural features indicate that the N1pC/P60 enzymes contain a fold similar to the papain-like peptidases, transglutaminases and arylamine acetyltransferases.
Figures


References
-
- Moat AG, Foster JW, Eds . Microbial Physiology. 4th. New York: John Wiley; 2002.
-
- Madigan MM, Parker J, Madigan MT, Martinko JM. Brock Biology of Microorganisms. 10th. Upper Saddle River, NJ: Prentice Hall; 2002.
-
- van Heijenoort J. Formation of the glycan chains in the synthesis of bacterial peptidoglycan. Glycobiology. 2001;11:25R–36R. - PubMed
-
- Healy VL, Lessard IA, Roper DI, Knox JR, Walsh CT. Vancomycin resistance in enterococci: reprogramming of the D-Ala-D-Ala ligases in bacterial peptidoglycan biosynthesis. Chem Biol. 2000;7:R109–R119. - PubMed
-
- Born TL, Blanchard JS. Structure/function studies on enzymes in the diaminopimelate pathway of bacterial cell wall biosynthesis. Curr Opin Chem Biol. 1999;3:607–613. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

