Lateral gene transfer and ancient paralogy of operons containing redundant copies of tryptophan-pathway genes in Xylella species and in heterocystous cyanobacteria
- PMID: 12620124
- PMCID: PMC151304
- DOI: 10.1186/gb-2003-4-2-r14
Lateral gene transfer and ancient paralogy of operons containing redundant copies of tryptophan-pathway genes in Xylella species and in heterocystous cyanobacteria
Abstract
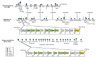
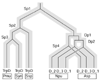
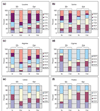
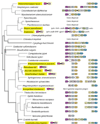
Background: Tryptophan-pathway genes that exist within an apparent operon-like organization were evaluated as examples of multi-genic genomic regions that contain phylogenetically incongruous genes and coexist with genes outside the operon that are congruous. A seven-gene cluster in Xylella fastidiosa includes genes encoding the two subunits of anthranilate synthase, an aryl-CoA synthetase, and trpR. A second gene block, present in the Anabaena/Nostoc lineage, but not in other cyanobacteria, contains a near-complete tryptophan operon nested within an apparent supraoperon containing other aromatic-pathway genes.
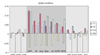
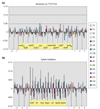
Results: The gene block in X. fastidiosa exhibits a sharply delineated low-GC content. This, as well as bias of codon usage and 3:1 dinucleotide analysis, strongly implicates lateral gene transfer (LGT). In contrast, parametric studies and protein tree phylogenies did not support the origination of the Anabaena/Nostoc gene block by LGT.
Conclusions: Judging from the apparent minimal amelioration, the low-GC gene block in X. fastidiosa probably originated by LGT at a relatively recent time. The surprising inability to pinpoint a donor lineage still leaves room for alternative, albeit less likely, explanations other than LGT. On the other hand, the large Anabaena/Nostoc gene block does not seem to have arisen by LGT. We suggest that the contemporary Anabaena/Nostoc array of divergent paralogs represents an ancient ancestral state of paralog divergence, with extensive streamlining by gene loss occurring in the lineage of descent representing other (unicellular) cyanobacteria.
Figures


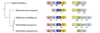

References
-
- Margulis L. Symbiosis in Cell Evolution. San Francisco: WH Freeman; 1981.
-
- Gray MW. Evolution of organellar genomes. Curr Opin Genet Dev. 1999;9:678–687. - PubMed
-
- Aravind L, Tatusov RL, Wolf YI, Walker DR, Koonin EV. Evidence for massive gene exchange between archaeal and bacterial hyperthermophiles. Trends Genet. 1998;14:442–444. - PubMed
-
- Nelson KE, Clayton RA, Gill SR, Gwinn ML, Dodson RJ, Haft DH, Hickey EK, Peterson JD, Nelson WC, Ketchum KA, et al. Evidence for lateral gene transfer between archaea and bacteria from genome sequence of Thermotoga maritima. Nature. 1999;399:323–329. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

