Isolation of novel ultramicrobacteria classified as actinobacteria from five freshwater habitats in Europe and Asia
- PMID: 12620827
- PMCID: PMC150105
- DOI: 10.1128/AEM.69.3.1442-1451.2003
Isolation of novel ultramicrobacteria classified as actinobacteria from five freshwater habitats in Europe and Asia
Abstract
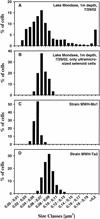
We describe the first freshwater members of the class Actinobacteria that have been isolated. Nine ultramicro-size (<0.1 microm(3)) strains were isolated from five freshwater habitats in Europe and Asia. These habitats represent a broad spectrum of ecosystems, ranging from deep oligotrophic lakes to shallow hypertrophic lakes. Even when the isolated strains were grown in very rich media, the cell size was <0.1 microm(3) and was indistinguishable from the cell sizes of bacteria belonging to the smaller size classes of natural lake bacterioplankton. Hybridization of the isolates with oligonucleotide probes and phylogenetic analysis of the 16S rRNA gene sequences of the isolated strains revealed that they are affiliated with the class Actinobacteria and the family Microbacteriaceae. The previously described species with the highest levels of sequence similarity are Clavibacter michiganensis and Rathayibacter tritici, two phytopathogens of terrestrial plants. The 16S rRNA gene sequences of the nine isolates examined are more closely related to cloned sequences from uncultured freshwater bacteria than to the sequences of any previously isolated bacteria. The nine ultramicrobacteria isolated form, together with several uncultured bacteria, a diverse phylogenetic cluster (Luna cluster) consisting exclusively of freshwater bacteria. Isolates obtained from lakes that are ecologically different and geographically separated by great distances possess identical 16S rRNA gene sequences but have clearly different ecophysiological and phenotypic traits. Predator-prey experiments demonstrated that at least one of the ultramicro-size isolates is protected against predation by the bacterivorous nanoflagellate Ochromonas sp. strain DS.
Figures



References
-
- Andersson, A., U. Larsson, and Å. Hagström. 1986. Size-selective grazing by a microflagellate on pelagic bacteria. Mar. Ecol. Prog. Ser. 33:51-57.
-
- Benlloch, S., A. Lopez-Lopez, E. O. Casamayor, L. Ovreas, V. Goddard, F. L. Daae, G. Smerdon, R. Massana, I. Jiont, F. Thingstad, C. Pedros-Alio, and F. Rodriguez-Valera. 2002. Prokaryotic genetic diversity throughout the salinity gradient of a coastal solar saltern. Environ. Microbiol. 4:349-360. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

