Comparative genomic analyses of the vibrio pathogenicity island and cholera toxin prophage regions in nonepidemic serogroup strains of Vibrio cholerae
- PMID: 12620865
- PMCID: PMC150053
- DOI: 10.1128/AEM.69.3.1728-1738.2003
Comparative genomic analyses of the vibrio pathogenicity island and cholera toxin prophage regions in nonepidemic serogroup strains of Vibrio cholerae
Abstract
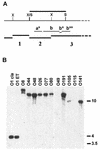
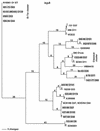
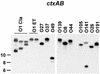
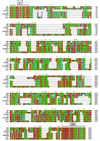
Two major virulence factors are associated with epidemic strains (O1 and O139 serogroups) of Vibrio cholerae: cholera toxin encoded by the ctxAB genes and toxin-coregulated pilus encoded by the tcpA gene. The ctx genes reside in the genome of a filamentous phage (CTXphi), and the tcpA gene resides in a vibrio pathogenicity island (VPI) which has also been proposed to be a filamentous phage designated VPIphi. In order to determine the prevalence of horizontal transfer of VPI and CTXphi among nonepidemic (non-O1 and non-O139 serogroups) V. cholerae, 300 strains of both clinical and environmental origin were screened for the presence of tcpA and ctxAB. In this paper, we present the comparative genetic analyses of 11 nonepidemic serogroup strains which carry the VPI cluster. Seven of the 11 VPI(+) strains have also acquired the CTXphi. Multilocus sequence typing and restriction fragment length polymorphism analyses of the VPI and CTXphi prophage regions revealed that the non-O1 and non-O139 strains were genetically diverse and clustered in lineages distinct from that of the epidemic strains. The left end of the VPI in the non-O1 and non-O139 strains exhibited extensive DNA rearrangements. In addition, several CTXphi prophage types characterized by novel repressor (rstR) and ctxAB genes and VPIs with novel tcpA genes were found in these strains. These data suggest that the potentially pathogenic, nonepidemic, non-O1 and non-O139 strains identified in our study most likely evolved by sequential horizontal acquisition of the VPI and CTXphi independently rather than by exchange of O-antigen biosynthesis regions in an existing epidemic strain.
Figures




References
-
- Albert, M. J., M. Ansaruzzaman, P. K. Bardhan, A. S. G. Faruque, S. M. Faruque, M. S. Islam, D. Mahalanabis, R. B. Sack, M. A. Salam, A. K. Siddique, M. D. Yunus, and K. Zaman. 1993. Large epidemic of cholera-like disease in Bangladesh caused by Vibrio cholerae O139 synonym Bengal. Lancet 342:387-390. - PubMed
-
- Aldova, E., K. Laznickova, E. Stepankova, and J. Lietava. 1968. Isolation of nonagglutinable vibrios from an enteritis outbreak in Czechoslovakia. J. Infect. Dis. 118:25-31. - PubMed
-
- Berche, P., C. Poyart, E. Abachin, H. Lelievre, J. Vandepitte, A. Dodin, and J.-M. Fournier. 1994. The novel epidemic strain O139 is closely related to pandemic strain O1 of Vibrio cholerae. J. Infect. Dis. 170:701-704. - PubMed
-
- Bhattacharya, S. K., M. K. Bhattacharya, G. B. Nair, D. Dutta, A. Deb, T. Ramamurthy, S. Garg, P. K. Saha, P. Dutta, A. Moitra, B. K. Mandel, T. Shimada, Y. Takeda, and B. C. Deb. 1993. Clinical profile of acute diarrhoea cases infected with the new epidemic strain of V. cholerae O139: designation of disease as cholera. J. Infect. Dis. 27:11-15. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

