In situ accessibility of small-subunit rRNA of members of the domains Bacteria, Archaea, and Eucarya to Cy3-labeled oligonucleotide probes
- PMID: 12620867
- PMCID: PMC150112
- DOI: 10.1128/AEM.69.3.1748-1758.2003
In situ accessibility of small-subunit rRNA of members of the domains Bacteria, Archaea, and Eucarya to Cy3-labeled oligonucleotide probes
Abstract
Low accessibility of the rRNA is together with cell wall impermeability and low cellular ribosome content a frequent reason for failure of whole-cell fluorescence hybridization with fluorescently labeled oligonucleotide probes. In this study we compare accessibility data for the 16S rRNA of Escherichia coli (gamma Proteobacteria, Bacteria) with the phylogenetically distantly related organisms Pirellula sp. strain 1 (Planctomycetes, Bacteria) and Metallosphaera sedula (Crenarchaeota, Archaea) and the 18S rRNA accessibility of Saccharomyces cerevisiae (Eucarya). For a total of 537 Cy3-labeled probes, the signal intensities of hybridized cells were quantified under standardized conditions by flow cytometry. The relative probe-conferred fluorescence intensities are shown on color-coded small-subunit rRNA secondary-structure models. For Pirellula sp., most of the probes belong to class II and III (72% of the whole data set), whereas most of the probes targeting sites on M. sedula were grouped into class V and VI (46% of the whole data set). For E. coli, 45% of all probes of the data set belong to class III and IV. A consensus model for the accessibility of the small-subunit rRNA to oligonucleotide probes is proposed which uses 60 homolog target sites of the three prokaryotic 16S rRNA molecules. In general, open regions were localized around helices 13 and 14 including target positions 285 to 338, whereas helix 22 (positions 585 to 656) and the 3' half of helix 47 (positions 1320 to 1345) were generally inaccessible. Finally, the 16S rRNA consensus model was compared to data on the in situ accessibility of the 18S rRNA of S. cerevisiae.
Figures
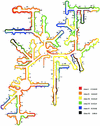
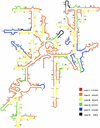
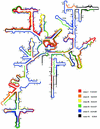


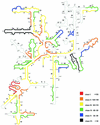

References
-
- Ban, N., P. Nissen, J. Hansen, M. Capel, P. B. Moore, and T. A. Steitz. 1999. Placement of protein and RNA structures into a 5 Å-resolution map of the 50S ribosomal subunit. Nature 400:841-847. - PubMed
-
- Brosius, J., T. J. Dull, D. D. Sleeter, and H. F. Noller. 1981. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J. Mol. Biol. 148:107-127. - PubMed
-
- Cai, J., I. N. Roberts, and M. D. Collins. 1996. Phylogenetic relationships among members of the Ascomycetous yeast genera Brettanomyces, Debaryomyces, Dekkera, and Kluyveromyces deduced by small-subunit rRNA gene sequences. Int. J. Syst. Bacteriol. 46:525-549. - PubMed
-
- Cannone, J. J., S. Subramanian, M. N. Schnare, J. R. Collett, L. M. D'Souza, Y. Du, B. Feng, N. Lin, L. V. Madabusi, K. M. Muller, N. Pande, Z. Shang, N. Yu, and R. R. Gutell. 2002. The Comparative RNA Web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics 3:15. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

